
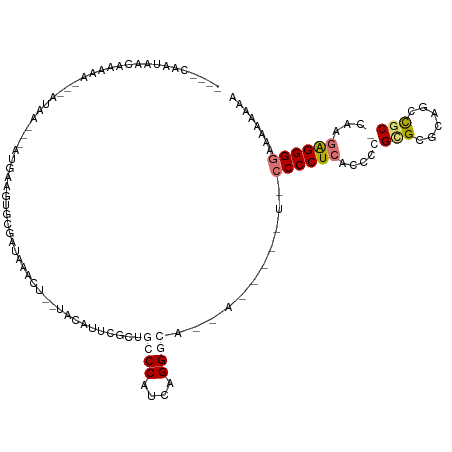
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,199,419 – 2,199,530 |
| Length | 111 |
| Max. P | 0.945747 |

| Location | 2,199,419 – 2,199,528 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 55.90 |
| Shannon entropy | 0.71015 |
| G+C content | 0.49745 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -11.64 |
| Energy contribution | -12.83 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2199419 109 - 21146708 GAACCAAUAACAAAAAGCGUUAAU---CGAAGUGCGGUGUAGC--UACGAUACCUGCCCUUCGGGCAACUU----AUUCCCCUCACCCCGCGCAGAGCCAC-UGAAGGGGGCAAUAAAA ................(((((..(---(((((.((((.(((..--.....))))))).))))))..)))..----..........((((...(((.....)-))..))))))....... ( -30.70, z-score = -1.12, R) >droSim1.chr2h_random 565685 98 - 3178526 ----CAAAAAAAAAAAGCAACAGUGGAUAAGGAGCGAUAAACU--UGGAUUCUCUGCCCAUUGGG-------------CCCCUCACCACGUG-GCAGCCGC-CAAGAGGGUCAGAAAAA ----................((((((.((..((((.(......--).).)))..)).))))))((-------------(((.((......((-((....))-)).)))))))....... ( -23.80, z-score = 0.13, R) >droYak2.chrU 26016091 105 + 28119190 ----CAAUAACAACAA--CAUAA---GUGUAGUGCGAUAGAC---UGCAUUUGUGGCCCGUCAGGGCAUCACU--GUCCCCCUCACACUGCGAGCAGCUGCACAAGAGGGGAAACAAAA ----............--(((((---(((((((.(....)))---))))))))))((((....)))).....(--((.((((((....((((......))))...))))))..)))... ( -37.20, z-score = -2.96, R) >droMoj3.scaffold_6498 1458798 108 + 3408170 -----AGCAACGAAAA---AUAA---AGGGAGUGCGACAAACUAAUACAUUCGUUGCCCAUCAGGGCCAAACUUAUUACCCCUCUCUGUGCGCGCCACUGCACAAGAGGGGAAAGAAAC -----.((((((((..---....---....(((.......)))......))))))))((....)).............(((((((.((((((......)))))))))))))........ ( -31.79, z-score = -2.49, R) >consensus ____CAAUAACAAAAA___AUAA___AUGAAGUGCGAUAAACU__UACAUUCGCUGCCCAUCAGGGCA__A_____U_CCCCUCACCCCGCGCGCAGCCGC_CAAGAGGGGAAAAAAAA .......................................................((((....))))...........((((((.....(((......)))....))))))........ (-11.64 = -12.83 + 1.19)


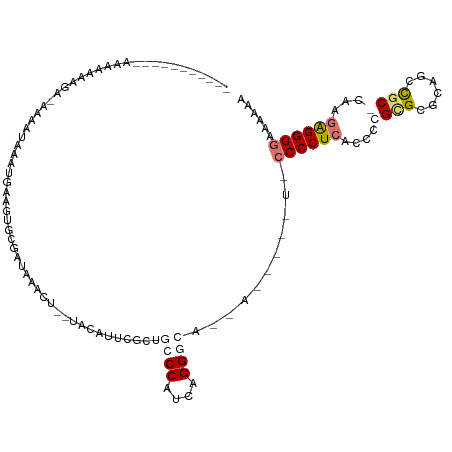
| Location | 2,199,421 – 2,199,530 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 51.63 |
| Shannon entropy | 0.77793 |
| G+C content | 0.50236 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -11.64 |
| Energy contribution | -12.83 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2199421 109 - 21146708 AUGAACCAAUAACAAAAAGCGUUAAU---CGAAGUGCGGUGUAGC--UACGAUACCUGCCCUUCGGGCAACUU----AUUCCCCUCACCCCGCGCAGAGCCAC-UGAAGGGGGCAAUAA ..................(((((..(---(((((.((((.(((..--.....))))))).))))))..)))..----..........((((...(((.....)-))..))))))..... ( -30.70, z-score = -0.88, R) >droSim1.chr2h_random 565687 96 - 3178526 ------CAAAAAAAAAAAGCAACAGUGGAUAAGGAGCGAUAAACU--UGGAUUCUCUGCCCAUUGGG-------------CCCCUCACCACGUG-GCAGCCGC-CAAGAGGGUCAGAAA ------................((((((.((..((((.(......--).).)))..)).))))))((-------------(((.((......((-((....))-)).)))))))..... ( -23.80, z-score = 0.20, R) >droYak2.chrU 26016093 103 + 28119190 -----------CAAUAACAACAACAUAAGUGUAGUGCGAUAGAC---UGCAUUUGUGGCCCGUCAGGGCAUCACU--GUCCCCCUCACACUGCGAGCAGCUGCACAAGAGGGGAAACAA -----------............((((((((((((.(....)))---))))))))))((((....)))).....(--((.((((((....((((......))))...))))))..))). ( -37.20, z-score = -2.86, R) >droMoj3.scaffold_6498 1458800 106 + 3408170 ------------AGCAACGA-AAAAUAAAGGGAGUGCGACAAACUAAUACAUUCGUUGCCCAUCAGGGCCAAACUUAUUACCCCUCUCUGUGCGCGCCACUGCACAAGAGGGGAAAGAA ------------.(((((((-(..........(((.......)))......))))))))((....)).............(((((((.((((((......)))))))))))))...... ( -31.79, z-score = -2.24, R) >consensus ___________AAAAAAAGA_AAAAUAAAUGAAGUGCGAUAAACU__UACAUUCGCUGCCCAUCAGGGCA__A_____U_CCCCUCACCCCGCGCGCAGCCGC_CAAGAGGGGAAAAAA .........................................................((((....))))...........((((((.....(((......)))....))))))...... (-11.64 = -12.83 + 1.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:30 2011