
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,146,271 – 2,146,415 |
| Length | 144 |
| Max. P | 0.980575 |

| Location | 2,146,271 – 2,146,415 |
|---|---|
| Length | 144 |
| Sequences | 5 |
| Columns | 153 |
| Reading direction | forward |
| Mean pairwise identity | 64.17 |
| Shannon entropy | 0.64151 |
| G+C content | 0.49180 |
| Mean single sequence MFE | -52.08 |
| Consensus MFE | -20.45 |
| Energy contribution | -21.93 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
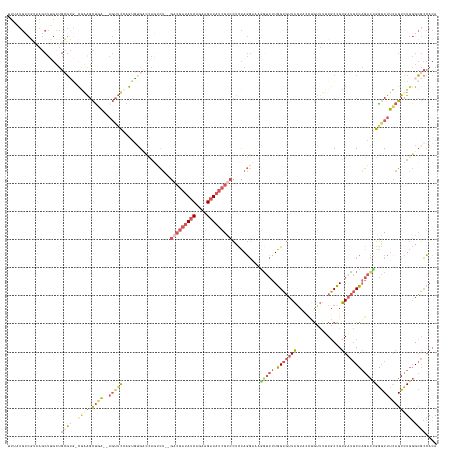
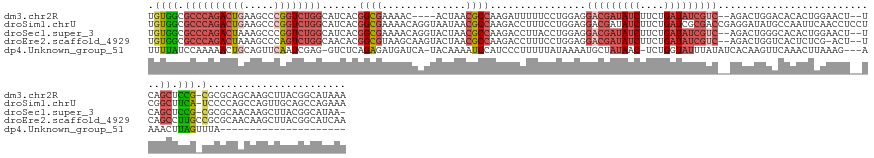
>dm3.chr2R 2146271 144 + 21146708 UUUAUGCCGUAAGCUUGCUGCGCG-CGGAGCUGA--AGUUCCAGUGUGUCCAGUCU--GACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAAAUCUUGGCGUUAGU----GUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACA .....((((((.(....))))).)-)(((((...--.))))).((((((((((.((--((.((((((....))))))((((...((((....))))(((((((((----(....)))..))))))).....)))).)))).))))))).))). ( -55.40, z-score = -2.65, R) >droSim1.chrU 1857335 152 + 15797150 UUUCUGGCUGCAACUGGCUGGGGA-UGAAGCCGAGGAGGUUGAAUUGGCAUAUCCUCGGUCGCGAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAUUACCUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACA ....((((.((..(((((((((((-(((.((((((((.(((.....)))...))))))))....(((....))).)))))).)))......(((.(((((..(........)..))))).)))))))).((((((....)))))))))))).. ( -62.20, z-score = -2.05, R) >droSec1.super_3 6919830 147 + 7220098 -UUAUGCCGUAAGCUUGUUGCGCG-CGGAGCUGA--AGUUCCAGUGUGCCCAGUCU--GACGAUAUCAGAAGAUAUCGUCCUCCAGGUAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCUUUAGUCUGGGCGCCACA -....(((((((.....))))).)-)(((((...--.))))).((((((((((.((--(((((((((....)))))))))....((((..(((((.(((((((................))))))))))))..)))).)).))))))).))). ( -62.39, z-score = -3.96, R) >droEre2.scaffold_4929 21984477 148 + 26641161 UUGAUGCCGUAAGCUUGUUGCGCGGCAAGGCUGA--AGU-CGAGAGUGACCAGUCU--GACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGCUUACGCCGUGUUGCCAGACUGGGCUUUAGUCUGGGCGCCACA ....((((((..((.....)))))))).(((...--.((-(.(((.((((((((((--(((((((((....)))))))))..........(((..((((((.(((....))).))))))....))))))))))...))).))).))))))... ( -60.80, z-score = -2.99, R) >dp4.Unknown_group_51 38428 126 - 55748 ---------------------UAAACUAAGUUUU---CUUUAAGUUUGAACUUGUGAUAUAAAUACCAGA-GUUAUAGCAUUUUAUAAAAAGGGAUGGAUUUUGUA-UGAUCAUCUCUGAGAC-CUCGAUUGAACUGCAGUUUUUGGAUAAAA ---------------------.....(((((((.---..........)))))))...........(((((-(.....(((.((((.....(((((((.(((.....-.))))))))))((...-.))...)))).)))...))))))...... ( -19.60, z-score = 0.86, R) >consensus UUUAUGCCGUAAGCUUGCUGCGCG_CGAAGCUGA__AGUUCCAGUGUGACCAGUCU__GACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACA ..........................((((((..........................(((((((((....)))))))))..........(((((.(((((((................)))))))))))).))))))............... (-20.45 = -21.93 + 1.48)


| Location | 2,146,271 – 2,146,415 |
|---|---|
| Length | 144 |
| Sequences | 5 |
| Columns | 153 |
| Reading direction | reverse |
| Mean pairwise identity | 64.17 |
| Shannon entropy | 0.64151 |
| G+C content | 0.49180 |
| Mean single sequence MFE | -44.73 |
| Consensus MFE | -17.62 |
| Energy contribution | -21.06 |
| Covariance contribution | 3.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
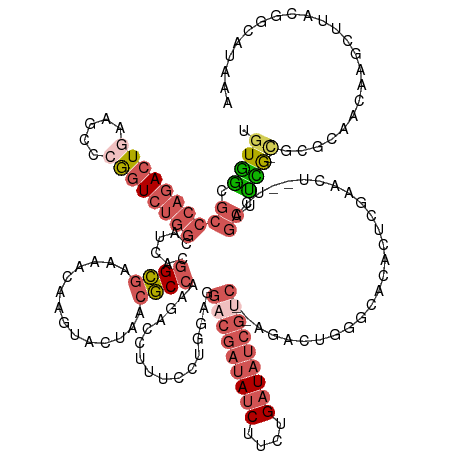
>dm3.chr2R 2146271 144 - 21146708 UGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAAC----ACUAACGCCAAGAUUUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUC--AGACUGGACACACUGGAACU--UCAGCUCCG-CGCGCAGCAAGCUUACGGCAUAAA (((.((((.(.((((((((.(((((.((.......((((.....----.....))))........(((.(...(((((((((....)))))))))--...).))))).)))))..))--)))).)).)-.)))).))).((.....))..... ( -51.40, z-score = -2.93, R) >droSim1.chrU 1857335 152 - 15797150 UGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAGGUAAUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUCGCGACCGAGGAUAUGCCAAUUCAACCUCCUCGGCUUCA-UCCCCAGCCAGUUGCAGCCAGAAA .(((..(((.((((((.....)))))))))..)))(((...(((.(((......)))..........((((.(((((..(((....)))..)).(((((((..((......))...))))))).....-)))))))...)))...)))..... ( -50.80, z-score = -1.68, R) >droSec1.super_3 6919830 147 - 7220098 UGUGGCGCCCAGACUAAAGCCCGGUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUACCUGGAGGACGAUAUCUUCUGAUAUCGUC--AGACUGGGCACACUGGAACU--UCAGCUCCG-CGCGCAACAAGCUUACGGCAUAA- (((.((((.(........((((((((((((.....((((..............))))....((((.....))))..((((((....)))))))))--))))))))).....(((.(.--...).))))-.)))).))).((.....))....- ( -54.04, z-score = -3.26, R) >droEre2.scaffold_4929 21984477 148 - 26641161 UGUGGCGCCCAGACUAAAGCCCAGUCUGGCAACACGGCGUAAGCAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUC--AGACUGGUCACUCUCG-ACU--UCAGCCUUGCCGCGCAACAAGCUUACGGCAUCAA .(((..(((.(((((.......))))))))..)))(((((.((......)).)))))..........(((((((((((((((....)))))))))--.((....)).......-.))--))))...(((((.((.....))...))))).... ( -52.20, z-score = -3.03, R) >dp4.Unknown_group_51 38428 126 + 55748 UUUUAUCCAAAAACUGCAGUUCAAUCGAG-GUCUCAGAGAUGAUCA-UACAAAAUCCAUCCCUUUUUAUAAAAUGCUAUAAC-UCUGGUAUUUAUAUCACAAGUUCAAACUUAAAG---AAAACUUAGUUUA--------------------- ..........((((((.((((...(((((-(((........)))).-.......................((((((((....-..))))))))...))..((((....))))...)---).)))))))))).--------------------- ( -15.20, z-score = -0.01, R) >consensus UGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUC__AGACUGGGCACACUCGAACU__UCAGCUUCG_CGCGCAACAAGCUUACGGCAUAAA .(((..(((.((((((.....)))))))))..)))((((..............))))................(((((((((....))))))))).......................................................... (-17.62 = -21.06 + 3.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:28 2011