| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,121,210 – 2,121,302 |
| Length | 92 |
| Max. P | 0.932866 |

| Location | 2,121,210 – 2,121,302 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.92 |
| Shannon entropy | 0.45212 |
| G+C content | 0.60142 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -20.86 |
| Energy contribution | -20.73 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
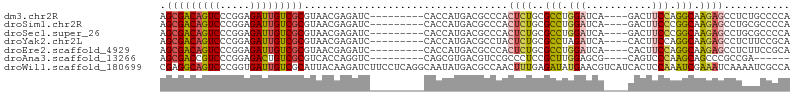
>dm3.chr2R 2121210 92 + 21146708 AGCGACAGUCCCGGAGAUUGUCGCGUAACGAGAUC---------CACCAUGACGCCCACUCUGCGCCUGGAUCA----GACUUCCAGGCAAGAGCCUCUGCCCCA .(((((((((.....))))))))).....(((...---------.....((.....))((((..(((((((...----....))))))).)))).)))....... ( -33.20, z-score = -2.63, R) >droSim1.chr2R 1018587 92 + 19596830 AGCGACAGUCCCGGAGAUUGUCGCGUAACGAGAUC---------CACCAUGACGCCCACUCUGCGCCUGGAUCA----GACUUCCCGGCAAGAGCCUGCGCCCCA .(((((((((.....)))))))))...........---------........(((...((((..(((.(((...----....))).))).))))...)))..... ( -31.30, z-score = -1.87, R) >droSec1.super_26 764112 92 + 826190 AGCGACAGUCCCGGAGAUUGUCGCGUAACGAGAUC---------CACCAUGACGCCCACUCUGCGCCUGGAUCA----GACUUCCCGGCAAGAGCCUGCGCCCCA .(((((((((.....)))))))))...........---------........(((...((((..(((.(((...----....))).))).))))...)))..... ( -31.30, z-score = -1.87, R) >droYak2.chr2L 1972180 92 - 22324452 AGCGACAGUCCCGGAGAUUGUCGCGUAACGAGAUC---------CACCAUGACGCCUACUCUGCGCCUAGAUCA----CACUUCCAGGCAAGAGCCUCUUCCGCA .(((((((((.....))))))))).....(((.((---------......))......((((..((((.((...----....)).)))).)))).)))....... ( -26.50, z-score = -1.30, R) >droEre2.scaffold_4929 6436101 92 - 26641161 AGCGACAGUCCCGGAGAUUGUCGCGUAACGAGAUC---------CACCAUGACGCCCACUCUGCGCCUGGAUCA----CACUUCCAGGCAAGAGCCUCUUCCGCA .(((((((((.....))))))))).....(((...---------.....((.....))((((..(((((((...----....))))))).)))).)))....... ( -33.20, z-score = -2.84, R) >droAna3.scaffold_13266 1202197 86 - 19884421 AGCGACCGUCCCGGAGACUGUCGCGUCACCAGGUC---------CAGCGUGACGUCCGCCCUCCGCUUGGAGCG----CAGUCCCAAGCAGCCCGCCGA------ .(((........((((..((..((((((((.....---------..).))))))).))..))))((((((..(.----..)..))))))....)))...------ ( -29.60, z-score = -0.21, R) >droWil1.scaffold_180699 851002 105 - 2593675 CGAGGCAGUCCCGGUGAUUGUCGCAUUACAAGAUCUUCCUCAGGCAAUAUGACGCCAACUUUGAGAUAUGAACGUCAUCACUCCAAAUCGAAAUCAAAAUCGCCA .((((.((((...(((((......)))))..)).)).)))).(((...((((((..................))))))...........((........))))). ( -19.77, z-score = 0.45, R) >consensus AGCGACAGUCCCGGAGAUUGUCGCGUAACGAGAUC_________CACCAUGACGCCCACUCUGCGCCUGGAUCA____CACUUCCAGGCAAGAGCCUCUGCCCCA .(((((((((.....)))))))))..................................((((..(((.(((...........))).))).))))........... (-20.86 = -20.73 + -0.14)

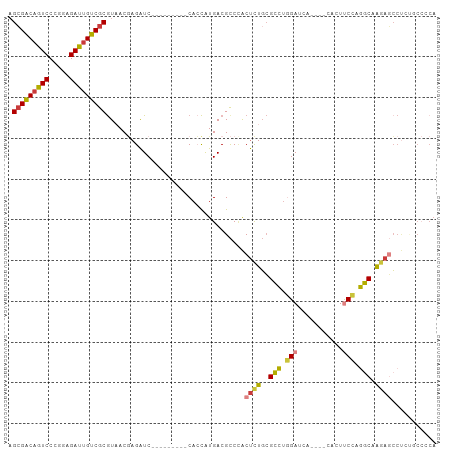
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:26 2011