| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,115,330 – 2,115,497 |
| Length | 167 |
| Max. P | 0.797112 |

| Location | 2,115,330 – 2,115,497 |
|---|---|
| Length | 167 |
| Sequences | 3 |
| Columns | 167 |
| Reading direction | forward |
| Mean pairwise identity | 77.82 |
| Shannon entropy | 0.30989 |
| G+C content | 0.32552 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -17.80 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2115330 167 + 21146708 AUUAACAGCAAUCAUAGUUAGUAAGUGAAAAUAUGAAAUCGAUAGGGUUCGGUUGAGUAGGAGUUUGAUGUAGGUUACCAUCUAUUUUCAAGAAAGAAAAAAUAUACAUAUGUAAAUUAUGUAUGCACUAUUGUGCUCUUUAAAAACAAAUACAAACCCAACGAAUC .....((((..(((((.(((.....)))...)))))...(((......))))))).((.((.(((((..((((((....)))))).......(((((......(((((((.......)))))))((((....)))))))))...........))))))).))..... ( -31.90, z-score = -1.11, R) >droSim1.chr2R 1012183 162 + 19596830 AUUGAUUCCAAUCAUAUUUAGUAAUUGAAAAUAUGAAAUCAAUAGGGUUCGGUUGGGUAGGAGUUUGGAUUAGGCUUCCAUCU----UCCAAGCCCAGUAUUUUCAAGAAAGUAGAA-ACAUAGAAACAUGUGGGCUAUAUAAAAUCCAAUACAAACCCAACGAAUU (((((((....((((((((.........))))))))))))))).(((((..(((((((.((((..((((.......))))..)----))).(((((..((((((....))))))...-((((......))))))))).......)))))))...)))))........ ( -45.00, z-score = -3.65, R) >droSec1.super_26 757949 154 + 826190 AUUGAUUCCAAUCAUAUUUAGUAAUUGAAAAUAUGAAAUCAAUAGGGUUCGGUUGAGUAGGAGUUUGGAUUAGGCUUCCAUCU----UCCAAGCCCAGUAUUUUCAAGAAAGUAGAA-ACAUA--------UGGGCUAUAUAAAAUCCAAUACAAACCCAACGAAUC (((((((....((((((((.........))))))))))))))).(((((..((((....((((..((((.......))))..)----))).((((((....((((.........)))-)....--------))))))..........))))...)))))........ ( -39.40, z-score = -3.12, R) >consensus AUUGAUUCCAAUCAUAUUUAGUAAUUGAAAAUAUGAAAUCAAUAGGGUUCGGUUGAGUAGGAGUUUGGAUUAGGCUUCCAUCU____UCCAAGCCCAGUAUUUUCAAGAAAGUAGAA_ACAUA___AC___UGGGCUAUAUAAAAUCCAAUACAAACCCAACGAAUC (((((((....((((((((.........))))))))))))))).(((((..((((....((((((((...)))))).))............((((((.((((((....))))))(....)...........))))))..........))))...)))))........ (-17.80 = -18.70 + 0.90)

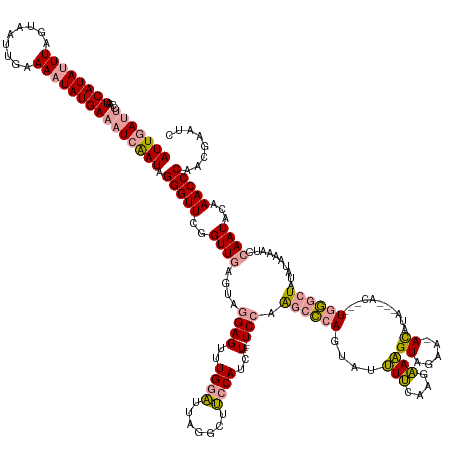
| Location | 2,115,330 – 2,115,497 |
|---|---|
| Length | 167 |
| Sequences | 3 |
| Columns | 167 |
| Reading direction | reverse |
| Mean pairwise identity | 77.82 |
| Shannon entropy | 0.30989 |
| G+C content | 0.32552 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -19.63 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2115330 167 - 21146708 GAUUCGUUGGGUUUGUAUUUGUUUUUAAAGAGCACAAUAGUGCAUACAUAAUUUACAUAUGUAUAUUUUUUCUUUCUUGAAAAUAGAUGGUAACCUACAUCAAACUCCUACUCAACCGAACCCUAUCGAUUUCAUAUUUUCACUUACUAACUAUGAUUGCUGUUAAU ((.(((..(((((((....(((((((((.((((((....))))(((((((.......)))))))..........)))))))))))((((........))))...............)))))))...)))..))..............((((..........)))).. ( -33.50, z-score = -2.80, R) >droSim1.chr2R 1012183 162 - 19596830 AAUUCGUUGGGUUUGUAUUGGAUUUUAUAUAGCCCACAUGUUUCUAUGU-UUCUACUUUCUUGAAAAUACUGGGCUUGGA----AGAUGGAAGCCUAAUCCAAACUCCUACCCAACCGAACCCUAUUGAUUUCAUAUUUUCAAUUACUAAAUAUGAUUGGAAUCAAU ........(((((((..((((.........((((((....((((.....-............))))....)))))).(((----...((((.......))))...)))...)))).))))))).(((((((((((((((.........))))))))....))))))) ( -37.43, z-score = -1.91, R) >droSec1.super_26 757949 154 - 826190 GAUUCGUUGGGUUUGUAUUGGAUUUUAUAUAGCCCA--------UAUGU-UUCUACUUUCUUGAAAAUACUGGGCUUGGA----AGAUGGAAGCCUAAUCCAAACUCCUACUCAACCGAACCCUAUUGAUUUCAUAUUUUCAAUUACUAAAUAUGAUUGGAAUCAAU (((((...(((((((..((((.........((((((--------....(-(((.........))))....)))))).(((----...((((.......))))...)))...)))).)))))))........((((((((.........))))))))...)))))... ( -37.60, z-score = -2.51, R) >consensus GAUUCGUUGGGUUUGUAUUGGAUUUUAUAUAGCCCA___GU___UAUGU_UUCUACUUUCUUGAAAAUACUGGGCUUGGA____AGAUGGAAGCCUAAUCCAAACUCCUACUCAACCGAACCCUAUUGAUUUCAUAUUUUCAAUUACUAAAUAUGAUUGGAAUCAAU ..((((((((((..((.(((((((.......(((((..................................))))).............((...)).)))))))))....)))))).))))....(((((((((((((((.........))))))))....))))))) (-19.63 = -19.98 + 0.35)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:25 2011