
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,109,030 – 2,109,135 |
| Length | 105 |
| Max. P | 0.737973 |

| Location | 2,109,030 – 2,109,135 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 56.49 |
| Shannon entropy | 0.93335 |
| G+C content | 0.44801 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -1.09 |
| Energy contribution | -1.02 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.04 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.737973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2109030 105 - 21146708 -UUUUA---UUUCUCGCACAGUGGUUCGCUAUUUCGCGGAACGUGC-AAACUUGGGAAGAUACACUUAUAUUUUUCACUAUUGAACCA--UUUUGCCACACGGUUCGCUUCU----- -.....---......((((.((..(((((......)))))))))))-.......((((((((......))))))))......(((((.--...........)))))......----- ( -21.00, z-score = -1.10, R) >droSim1.chr2R 1005490 105 - 19596830 -UUUUA---UUUCUCGCACAGUGGUUCGCUACUUUGCGGAACAUGC-GAACUUGGGCAGAUACACUUAUAUUUUUCACUAUUGAACCA--UUUUGCCACACGGUUCGCUUCA----- -.....---.(((.((((.(((((....))))).)))))))...((-(((((((((((((.......(((........))).......--.)))))).)).)))))))....----- ( -30.56, z-score = -4.75, R) >droSec1.super_26 751147 105 - 826190 -UUUUA---UUUCUCGCACAGUGGUUCGCUACUUUGCGGAACAUGC-GAACUUGGGCAGAUACACUUAUAUUUUUCACUAUUGAACCA--UUUUGCCACACGGUUCGCUUCA----- -.....---.(((.((((.(((((....))))).)))))))...((-(((((((((((((.......(((........))).......--.)))))).)).)))))))....----- ( -30.56, z-score = -4.75, R) >droYak2.chr2L 1960871 105 + 22324452 -UUUUA---UUUCUCGCAUUGUGGUUCGCUAUUUUGCGGAACGUGC-GAACUUGGCCAGGCACAUUAAUUUUUUGCACUUUUAAACAA--UUUUGUCACACUGUUCGCUUCU----- -.....---.(((.((((..((((....))))..)))))))...((-((((.((....((((....((((.((((......)))).))--)).)))).))..))))))....----- ( -22.40, z-score = -1.20, R) >droEre2.scaffold_4929 6421694 105 + 26641161 -UUUUA---UUUCUCGCACAGUGGUUCGCUCUUUCGCGGAACGUGC-GAACUUGGGCAGGCACAUUAAUUGUUUGCACUUUUGAACAA--UUUUGCCACACUGUUCGCUUCU----- -.....---.....((.((((((....((((.((((((.....)))-)))...)))).((((....((((((((........))))))--)).)))).)))))).)).....----- ( -32.40, z-score = -3.22, R) >droAna3.scaffold_13266 1191018 105 + 19884421 -UUUUU---UUUAUCGCACAGUGGCACG-ACACCUACGAGUCGUGA-GCGCCUCUCUCUCAGCACCGAUUUGAGCCGUU-CCAGGCCACUCC-UGGCCCACUGUGCGCUCUCC---- -.....---.....((((((((((((((-((........))))))(-(((.(((..((........))...))).))))-(((((.....))-))).))))))))))......---- ( -38.30, z-score = -3.78, R) >dp4.chr3 18036600 110 - 19779522 -UUUUA---UUUCCCGCACAGUGGUUCACGCGCUCACGAAAUUCGACUCUCAUGGGAAUGAGCACCAAUUAGUCGCACU-UUCCA-CACUCU-CUGCACACUGUGCGCGCGCUCUCU -.....---.....(((((((((........(((((((.....)).(((....)))..)))))...........(((..-.....-......-.))).))))))))).......... ( -27.14, z-score = -1.11, R) >droWil1.scaffold_180699 817841 113 + 2593675 UUUUUAGAUUUUCCCGCACAGUGACACGACUUUGAAUGGUAAAUAC-UUUGUUUGG-AGGCGCA-AAAUUUUUCACACAAUUUUACUAGCUUUCGUAACACUGUGCACUGUGAUAU- ...............((((((((..((((.......(((((((...-.((((.(((-(((....-....)))))).)))).)))))))....))))..))))))))..........- ( -27.90, z-score = -2.56, R) >droVir3.scaffold_12875 4868040 110 - 20611582 UUUAUU---UU-CACGCACAGUGUCACAAAAGCGCACAACAUGCCGUGCACAUGGCGCCAUACAACAUUUCAAG-CAUG-UUCAACAGCACUCGAGCACACUGUGCGUGCAAAUGU- ..((((---(.-((((((((((((.......((((((........))))..(((....)))............)-)..(-(((..........)))))))))))))))).))))).- ( -34.90, z-score = -2.16, R) >droMoj3.scaffold_6496 1657297 108 + 26866924 UUUAUU---UU-CACGCACAGUGGCACAAACGCGUCAUAAAUAUUCAGCACAUGGUG-UGCAAACCAAUUCAAG-CAUG-UUCACCAAUACU-AGUCACACAGUGCGUGCAAAUGC- ..((((---(.-(((((((.(((((......((..............))...(((((-.(((..(........)-..))-).))))).....-.)))))...))))))).))))).- ( -28.34, z-score = -1.93, R) >droGri2.scaffold_15245 18192489 110 + 18325388 UUUAUU---UUGCAUGCACAGUGGCGUAAGAGUGCGCCGCAUGCUUCUCCCAUGAGACCGCAUUGAAUUUCAGG-CAUU-UCGAACAGCACU-CGACACACUGUGCGCACAAUAAU- .(((((---.(((..((((((((..((..(((((((((..((((.((((....))))..))))((.....))))-)...-.......)))))-).)).))))))))))).))))).- ( -36.80, z-score = -2.83, R) >consensus _UUUUA___UUUCUCGCACAGUGGUUCGCUAGUUCGCGGAACGUGC_GAACUUGGGCAGGCACACUAAUUUUUU_CACU_UUGAACAA__UUUUGCCACACUGUGCGCUCCA_____ ..............((.((.(((...........................................................................))).)).)).......... ( -1.09 = -1.02 + -0.06)
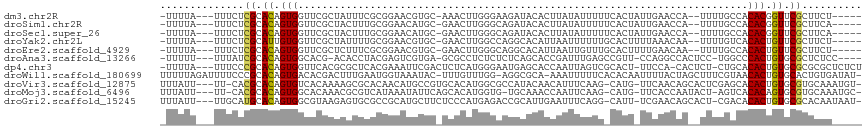


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:24 2011