| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,098,567 – 2,098,658 |
| Length | 91 |
| Max. P | 0.987883 |

| Location | 2,098,567 – 2,098,658 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 56.51 |
| Shannon entropy | 0.85950 |
| G+C content | 0.34102 |
| Mean single sequence MFE | -16.16 |
| Consensus MFE | -4.14 |
| Energy contribution | -6.27 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.882049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
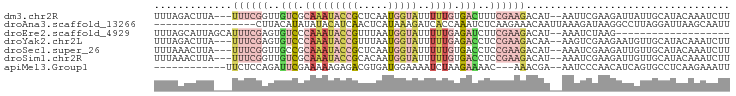
>dm3.chr2R 2098567 91 + 21146708 UUUAGACUUA---UUUCGGUUGUCGCAAAUACCGCUCAAUGGUAUUUUUGUGACUUUCGAAGACAU--AAUUCGAAGAUUAUUGCAUACAAAUCUU ....((.(((---((((((..(((((((((((((.....))))))..)))))))..)))))...))--)).)).((((((..........)))))) ( -21.60, z-score = -2.77, R) >droAna3.scaffold_13266 1178519 79 - 19884421 -----------------CUUACAUAUAUACAUCAACUCAUAAAGAUCACCAAAUCUCAAGAAACAAUUAAAGAUAAGGCCUUAGGAUUAAGCAAUU -----------------..................................((((..(((...................)))..))))........ ( -2.91, z-score = 0.99, R) >droEre2.scaffold_4929 6410209 75 - 26641161 UUUAGCAUUAGCAUUUCGAGUGUCCCAAAUACCGUUUAAUGGUAUUUUUGAGAUCUUCGAAGACAU--AAAUCUAAG------------------- .((((..(((...(((((((.(((.((((((((((...)))))))..))).))).)))))))...)--))..)))).------------------- ( -17.00, z-score = -3.13, R) >droYak2.chr2L 1947644 91 - 22324452 UUUAGACUUA---UUUCGAGUGUCCCAAAUACCGUUUAAUGGUAUUUUUGAGACCUCCGAAGACAA--AAGUCGAAGAAUGUUGCAUACAAAUCUU ....(((((.---(((((((.(((.((((((((((...)))))))..))).))))).)))))....--))))).((((.(((.....)))..)))) ( -20.20, z-score = -2.46, R) >droSec1.super_26 741338 91 + 826190 UUUAAACUUA---UUUCGGUUGCCGCAAAUACCGCUCAAUGGUAUUUUUGUGACCUCCGAAGACAU--AAAUCGAAGAUUGUUGCAUACAAAUCUU ..........---((((((..(.(((((((((((.....))))))..))))).)..))))))....--......((((((((.....)).)))))) ( -19.50, z-score = -1.98, R) >droSim1.chr2R 994892 91 + 19596830 UUUAAACUUA---UUUCGGUUGUCGCAAAUACCGCACAAUGGUAUUUUUGUGACCUCCGAAGACAU--AAAUCGAAGAUUGUUGCAUACAAAUCUU ..........---((((((..(((((((((((((.....))))))..)))))))..))))))....--......((((((((.....)).)))))) ( -24.50, z-score = -3.65, R) >apiMel3.Group1 9441005 79 - 25854376 ------------UUCUCCAGAUUCGAAAAAGAGACGUGAUGGAAAAUCUAAGAAAAC---AAACGA--AAUCCCAACAUCAGUGCCUCAAGAAAUU ------------..................(((.(((((((................---......--........))))).)).)))........ ( -7.41, z-score = 0.84, R) >consensus UUUAGACUUA___UUUCGAUUGUCGCAAAUACCGCUUAAUGGUAUUUUUGAGACCUCCGAAGACAU__AAAUCGAAGAUUGUUGCAUACAAAUCUU .............((((((..(((.(((((((((.....)))))..)))).)))..)))))).................................. ( -4.14 = -6.27 + 2.13)
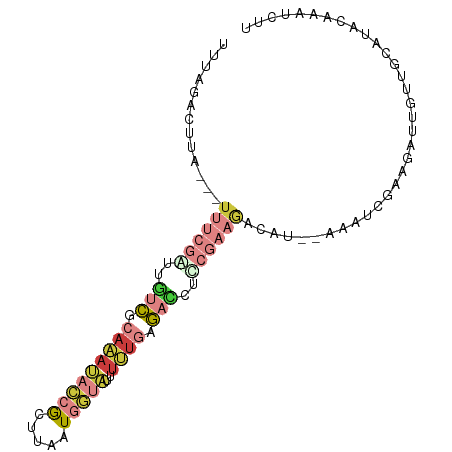

| Location | 2,098,567 – 2,098,658 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 56.51 |
| Shannon entropy | 0.85950 |
| G+C content | 0.34102 |
| Mean single sequence MFE | -18.40 |
| Consensus MFE | -6.58 |
| Energy contribution | -7.04 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.88 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.987883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
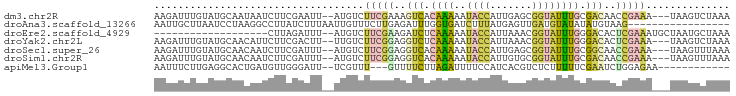
>dm3.chr2R 2098567 91 - 21146708 AAGAUUUGUAUGCAAUAAUCUUCGAAUU--AUGUCUUCGAAAGUCACAAAAAUACCAUUGAGCGGUAUUUGCGACAACCGAAA---UAAGUCUAAA .((((((((...........((((((..--.....)))))).(((.(((..(((((.......)))))))).))).......)---)))))))... ( -19.60, z-score = -2.33, R) >droAna3.scaffold_13266 1178519 79 + 19884421 AAUUGCUUAAUCCUAAGGCCUUAUCUUUAAUUGUUUCUUGAGAUUUGGUGAUCUUUAUGAGUUGAUGUAUAUAUGUAAG----------------- ...(((((((..(.((((((..(((((............)))))..)))...)))...)..)))).)))..........----------------- ( -9.70, z-score = 0.47, R) >droEre2.scaffold_4929 6410209 75 + 26641161 -------------------CUUAGAUUU--AUGUCUUCGAAGAUCUCAAAAAUACCAUUAAACGGUAUUUGGGACACUCGAAAUGCUAAUGCUAAA -------------------.((((..((--(.((.(((((...((((((..(((((.......)))))))))))...)))))..)))))..)))). ( -15.90, z-score = -2.67, R) >droYak2.chr2L 1947644 91 + 22324452 AAGAUUUGUAUGCAACAUUCUUCGACUU--UUGUCUUCGGAGGUCUCAAAAAUACCAUUAAACGGUAUUUGGGACACUCGAAA---UAAGUCUAAA .((((((((..............(((..--..)))(((((..(((((((..(((((.......))))))))))))..))))))---)))))))... ( -26.30, z-score = -4.17, R) >droSec1.super_26 741338 91 - 826190 AAGAUUUGUAUGCAACAAUCUUCGAUUU--AUGUCUUCGGAGGUCACAAAAAUACCAUUGAGCGGUAUUUGCGGCAACCGAAA---UAAGUUUAAA ((((((.((.....)))))))).(((((--((...(((((..(((.(((..(((((.......)))))))).)))..))))))---)))))).... ( -20.80, z-score = -1.58, R) >droSim1.chr2R 994892 91 - 19596830 AAGAUUUGUAUGCAACAAUCUUCGAUUU--AUGUCUUCGGAGGUCACAAAAAUACCAUUGUGCGGUAUUUGCGACAACCGAAA---UAAGUUUAAA ((((((.((.....)))))))).(((((--((...(((((..(((.(((..(((((.......)))))))).)))..))))))---)))))).... ( -21.40, z-score = -1.93, R) >apiMel3.Group1 9441005 79 + 25854376 AAUUUCUUGAGGCACUGAUGUUGGGAUU--UCGUUU---GUUUUCUUAGAUUUUCCAUCACGUCUCUUUUUCGAAUCUGGAGAA------------ ...(((((.(((....((((((((((..--((....---.........))..)))))..)))))((......)).))).)))))------------ ( -15.12, z-score = 0.05, R) >consensus AAGAUUUGUAUGCAACAAUCUUCGAUUU__AUGUCUUCGGAGGUCUCAAAAAUACCAUUAAGCGGUAUUUGCGACAACCGAAA___UAAGUCUAAA ...................................(((((..(((.((((..((((.......)))))))).)))..))))).............. ( -6.58 = -7.04 + 0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:20 2011