
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,077,883 – 2,077,986 |
| Length | 103 |
| Max. P | 0.979506 |

| Location | 2,077,883 – 2,077,986 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.21 |
| Shannon entropy | 0.55336 |
| G+C content | 0.50420 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -24.73 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
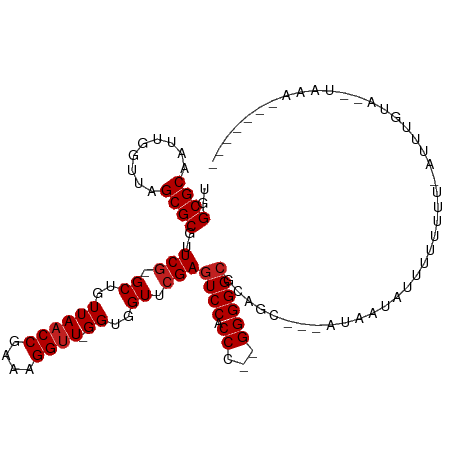
>dm3.chr2R 2077883 103 + 21146708 UGGCGCAAUUGGUUAGCGCGUUCG-GCUGUUAACCGAAAGGUU-GGUGGUUCGAGUCCACCC--GGGGGCGCAGCUACGUAAAAUUUUUUU--AUUUGUAAAUGAGAGG---- ..(((....(((((.((((.((((-(....(((((....))))-)((((.......))))))--))).))))))))))))......(((((--(((....)))))))).---- ( -33.40, z-score = -1.34, R) >droSim1.chr2R 973694 103 + 19596830 UGGCGCAAUUGGUUAGCGCGUUCG-GCUGUUAACCGAAAGGUU-GGUGGUUCGAGUCCACCC--GGGGGCGCAGCUUCCUAUUAUUUUUUUU-AUUCGUAA-UAAGAGG---- ..((((..((((...(.(..((((-((..((((((....))))-))..).)))))..).)))--))..))))..((((.((((((.......-....))))-)).))))---- ( -32.90, z-score = -1.19, R) >droSec1.super_26 720845 104 + 826190 UGGCGCAAUUGGUUAGCGCGUUCG-GCUGUUAACCGAAAGGUU-GGUGGUUCGAGUCCACCC--GGGGGCGCAGCUUCGUAUUAUUUUUUUUUAUUCGUAA-UAAGAGG---- ..((((..((((...(.(..((((-((..((((((....))))-))..).)))))..).)))--))..))))..((((.((((((............))))-)).))))---- ( -32.80, z-score = -1.31, R) >droYak2.chr2L 1925357 108 - 22324452 UGGCGCAAUUGGUUAGCGCGUUCG-GCUGUUAACCGAAAGGUU-GGUGGUUCGAGUCCACCC--GGGGGCGCAAAAUCAUAAUAUUUUUUUG-AUUUGUACCUUUAGGAAUGG ..((((.........)))).((((-((..((((((....))))-))..).))))).(((((.--.((((..((((.(((.((.....)).))-)))))..))))..))..))) ( -37.30, z-score = -2.38, R) >droEre2.scaffold_4929 6387270 111 - 26641161 UGGCGCAUUGCGGUAGCGCGUUCGCGCUGUUAUCCGAAAGGCU-GGGCGUUGGAGUCCACCCC-GGGGGGGGGCCGCACUUGUAUAUUUUUUAUUGCGCCCUUAAAUAAAAGG .((((((.(((((((((((....))))).....((....)).(-((((......)))))((((-....)))))))))).....(((.....)))))))))............. ( -43.40, z-score = -0.65, R) >dp4.chr3 1366063 91 - 19779522 UGGCGCAAUUGGUUAGCGCGUUCG-GCUGUUAACCGAAAGGUU-GGUGGUUCGAGUCCACCC--GGGGGCGCAUA--UAUC-UAUUUUUCCC-AUUUUU-------------- ..((((..((((...(.(..((((-((..((((((....))))-))..).)))))..).)))--))..))))...--....-..........-......-------------- ( -29.20, z-score = -0.90, R) >droPer1.super_73 214872 90 - 267513 UGGCGCAAUUGGUUAGCGCGUUCG-GCUGUUAACCGAAAGGUU-GGUGGUUCGAGUCCACCC--GGGGGCGCAUC----UAAUAUUUUACUU-CUCUUU-------------- .......(((((...((((.((((-(....(((((....))))-)((((.......))))))--))).))))..)----)))).........-......-------------- ( -29.50, z-score = -1.25, R) >droWil1.scaffold_180700 774150 88 - 6630534 UGGCGCAAUCGGUUAGCGCGUUCG-GCUGUUAACCGAAAGGUU-GGUGGUUCGAGUCCACCC--GGGGGCGCAAA---AUAACUUUUUUGUUCUU------------------ ..((((..((((...(.(..((((-((..((((((....))))-))..).)))))..).)))--))..))))...---.................------------------ ( -31.20, z-score = -1.51, R) >droVir3.scaffold_12875 15845008 103 - 20611582 UGGCGCAAUCGGUUAGCGCGUUCG-GCUGUUAACCGAAAGGUU-GGUGGUUCGAGUCCACCC--GGGGGCGUUUCA--AUAUUUUUUAUUUUUCUUCUUAUAUAAAUAG---- .(((((..((((...(.(..((((-((..((((((....))))-))..).)))))..).)))--))..)))))...--...............................---- ( -28.90, z-score = -1.39, R) >droMoj3.scaffold_6496 1613151 109 - 26866924 UGGCGCAAUCGGUUAGCGCGUUCG-GCUGUUAACCGAAAGGUUUGGUGGUUCGAGUCCACCCCGGGGGGCGCGGG---CAAACUUUUUUGCAAAUUCAUUUUUAUAUUACCCG ..((((..(((((((((((.....-)).)))))))))..((...(((((.......))))))).....))))(((---((((....)))))(((......))).......)). ( -34.60, z-score = -0.16, R) >droGri2.scaffold_14906 7468313 85 + 14172833 UGGCGCAAUUGGUUAGCGCGUUCG-GCUGUUAACCGAAAGGUU-GGUGGUUCGAGUCCACCC--GGGGGCGGA------UAUGAUUUUUUA--AUUU---------------- ..((((.........))))..(((-((..((((((....))))-))..).))))((((.((.--...)).)))------)...........--....---------------- ( -30.70, z-score = -2.09, R) >anoGam1.chr2L 3865486 99 - 48795086 UGGCGCAAUCGGUUAGCGCGUUCG-GCUGUUAACCGAAAGGUU-GGUGGUUCGAGUCCACCC--GGGGGCGCAGG---AAAUACUCUUU-------UAUUAUUAAAGAACAGG ..((((..((((...(.(..((((-((..((((((....))))-))..).)))))..).)))--))..))))...---......(((((-------.......)))))..... ( -32.80, z-score = -1.50, R) >consensus UGGCGCAAUUGGUUAGCGCGUUCG_GCUGUUAACCGAAAGGUU_GGUGGUUCGAGUCCACCC__GGGGGCGCAGC___AUAAUAUUUUUUUU_AUUUGUA__UAAA_______ ..((((..(((((((((((......)).))))))))).......(((((.......))))).......))))......................................... (-24.73 = -24.90 + 0.17)
| Location | 2,077,883 – 2,077,986 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.21 |
| Shannon entropy | 0.55336 |
| G+C content | 0.50420 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -16.37 |
| Energy contribution | -16.62 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2077883 103 - 21146708 ----CCUCUCAUUUACAAAU--AAAAAAAUUUUACGUAGCUGCGCCCCC--GGGUGGACUCGAACCACC-AACCUUUCGGUUAACAGC-CGAACGCGCUAACCAAUUGCGCCA ----................--............(((((..((((....--.(((((.......)))))-.....((((((.....))-)))).)))).......)))))... ( -22.70, z-score = -1.10, R) >droSim1.chr2R 973694 103 - 19596830 ----CCUCUUA-UUACGAAU-AAAAAAAAUAAUAGGAAGCUGCGCCCCC--GGGUGGACUCGAACCACC-AACCUUUCGGUUAACAGC-CGAACGCGCUAACCAAUUGCGCCA ----..(((((-(((.....-........))))))))....((((....--.(((((.......)))))-.....((((((.....))-)))).))))............... ( -24.82, z-score = -1.24, R) >droSec1.super_26 720845 104 - 826190 ----CCUCUUA-UUACGAAUAAAAAAAAAUAAUACGAAGCUGCGCCCCC--GGGUGGACUCGAACCACC-AACCUUUCGGUUAACAGC-CGAACGCGCUAACCAAUUGCGCCA ----.......-......................((..((((.......--.(((((.......)))))-((((....))))..))))-))...((((.........)))).. ( -21.50, z-score = -0.74, R) >droYak2.chr2L 1925357 108 + 22324452 CCAUUCCUAAAGGUACAAAU-CAAAAAAAUAUUAUGAUUUUGCGCCCCC--GGGUGGACUCGAACCACC-AACCUUUCGGUUAACAGC-CGAACGCGCUAACCAAUUGCGCCA ...........((..(((..-....(((((......)))))((((....--.(((((.......)))))-.....((((((.....))-)))).)))).......)))..)). ( -27.00, z-score = -2.01, R) >droEre2.scaffold_4929 6387270 111 + 26641161 CCUUUUAUUUAAGGGCGCAAUAAAAAAUAUACAAGUGCGGCCCCCCCCC-GGGGUGGACUCCAACGCCC-AGCCUUUCGGAUAACAGCGCGAACGCGCUACCGCAAUGCGCCA .............(((((((((.....))).....(((((((.......-.(((((........)))))-........)).....(((((....))))).))))).)))))). ( -36.09, z-score = -0.77, R) >dp4.chr3 1366063 91 + 19779522 --------------AAAAAU-GGGAAAAAUA-GAUA--UAUGCGCCCCC--GGGUGGACUCGAACCACC-AACCUUUCGGUUAACAGC-CGAACGCGCUAACCAAUUGCGCCA --------------......-(((....(((-....--)))...)))..--.(((((.......)))))-.....((((((.....))-)))).((((.........)))).. ( -24.50, z-score = -1.14, R) >droPer1.super_73 214872 90 + 267513 --------------AAAGAG-AAGUAAAAUAUUA----GAUGCGCCCCC--GGGUGGACUCGAACCACC-AACCUUUCGGUUAACAGC-CGAACGCGCUAACCAAUUGCGCCA --------------......-..((((....(((----(.((((.....--.(((((.......)))))-.....((((((.....))-))))))))))))....)))).... ( -23.60, z-score = -1.52, R) >droWil1.scaffold_180700 774150 88 + 6630534 ------------------AAGAACAAAAAAGUUAU---UUUGCGCCCCC--GGGUGGACUCGAACCACC-AACCUUUCGGUUAACAGC-CGAACGCGCUAACCGAUUGCGCCA ------------------.................---...((((....--.(((((.......)))))-......(((((((...((-.....))..)))))))..)))).. ( -23.60, z-score = -1.24, R) >droVir3.scaffold_12875 15845008 103 + 20611582 ----CUAUUUAUAUAAGAAGAAAAAUAAAAAAUAU--UGAAACGCCCCC--GGGUGGACUCGAACCACC-AACCUUUCGGUUAACAGC-CGAACGCGCUAACCGAUUGCGCCA ----..............................(--(((..((((...--.))))...))))......-.....((((((.....))-)))).((((.........)))).. ( -20.90, z-score = -1.13, R) >droMoj3.scaffold_6496 1613151 109 + 26866924 CGGGUAAUAUAAAAAUGAAUUUGCAAAAAAGUUUG---CCCGCGCCCCCCGGGGUGGACUCGAACCACCAAACCUUUCGGUUAACAGC-CGAACGCGCUAACCGAUUGCGCCA .((((((........((......)).......)))---)))((((....((((((((.......)))))......((((((.....))-))))........)))...)))).. ( -31.86, z-score = -0.80, R) >droGri2.scaffold_14906 7468313 85 - 14172833 ----------------AAAU--UAAAAAAUCAUA------UCCGCCCCC--GGGUGGACUCGAACCACC-AACCUUUCGGUUAACAGC-CGAACGCGCUAACCAAUUGCGCCA ----------------....--............------((((((...--.))))))...........-.....((((((.....))-)))).((((.........)))).. ( -23.10, z-score = -2.66, R) >anoGam1.chr2L 3865486 99 + 48795086 CCUGUUCUUUAAUAAUA-------AAAGAGUAUUU---CCUGCGCCCCC--GGGUGGACUCGAACCACC-AACCUUUCGGUUAACAGC-CGAACGCGCUAACCGAUUGCGCCA ....((((((.......-------)))))).....---...((((....--.(((((.......)))))-......(((((((...((-.....))..)))))))..)))).. ( -25.40, z-score = -1.45, R) >consensus _______UUUA__UACAAAU_AAAAAAAAUAAUAU___GCUGCGCCCCC__GGGUGGACUCGAACCACC_AACCUUUCGGUUAACAGC_CGAACGCGCUAACCAAUUGCGCCA ..........................................(((((....)))))...(((........((((....)))).......)))..((((.........)))).. (-16.37 = -16.62 + 0.25)
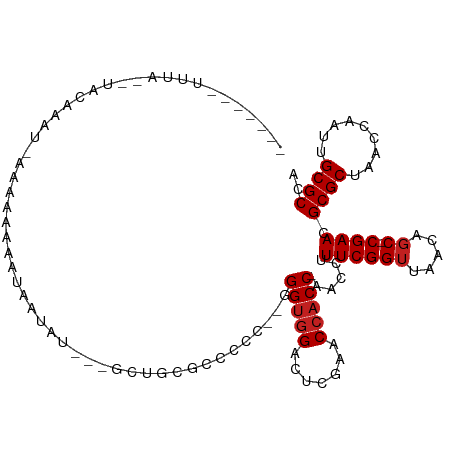
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:19 2011