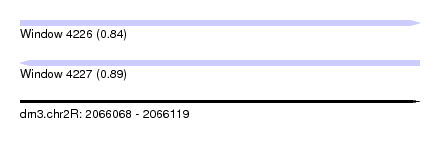
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,066,068 – 2,066,119 |
| Length | 51 |
| Max. P | 0.893930 |

| Location | 2,066,068 – 2,066,119 |
|---|---|
| Length | 51 |
| Sequences | 7 |
| Columns | 55 |
| Reading direction | forward |
| Mean pairwise identity | 78.82 |
| Shannon entropy | 0.43017 |
| G+C content | 0.39695 |
| Mean single sequence MFE | -8.17 |
| Consensus MFE | -8.10 |
| Energy contribution | -8.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2066068 51 + 21146708 --AAAAUAAAAAAAUUC--GAAUACGAUCCUGCCAGGAGUCGAACCUGGAAUCUU --...............--......(((....(((((.......))))).))).. ( -8.10, z-score = -0.90, R) >droYak2.chr2L 1912626 53 - 22324452 --AAAACAAAAAAGUGUAAAAAUUCGAUCCUGCCAGGAGUCGAACCUGGAAUCUU --.......................(((....(((((.......))))).))).. ( -8.10, z-score = -0.80, R) >droEre2.scaffold_4929 6374984 51 - 26641161 --AAAACAAAAAAGUG--AAAGUCCGAUCCUGCCAGGAGUCGAACCUGGAAUCUU --..............--.......(((....(((((.......))))).))).. ( -8.10, z-score = -0.38, R) >droPer1.super_73 202093 50 - 267513 --CAAAUAAAAAAGAU---AUCUCCGAUCCUGCCAGGAGUCGAACCUGGAAUCUU --..............---......(((....(((((.......))))).))).. ( -8.10, z-score = -0.55, R) >droWil1.scaffold_180700 756544 54 - 6630534 AAUGUAA-AAAAAUUAUACAUCUGCGAUCCUGCCAGGAGUCGAACCUGGAAUCUU .(((((.-........)))))....(((....(((((.......))))).))).. ( -8.60, z-score = -0.77, R) >droVir3.scaffold_12875 4798489 52 + 20611582 ---GUAAUGAAAAGAAAAAAUUAACGAUCCUGCCAGGAGUCGAACCUGGAAUCUU ---......................(((....(((((.......))))).))).. ( -8.10, z-score = -0.91, R) >droMoj3.scaffold_6496 1594950 52 - 26866924 ---AUAAUAAAAAGUUGAAUCUUGCGAUCCUGCCAGGAGUCGAACCUGGAAUCUU ---......................(((....(((((.......))))).))).. ( -8.10, z-score = -0.18, R) >consensus __AAAAAAAAAAAGUU_AAAAAUACGAUCCUGCCAGGAGUCGAACCUGGAAUCUU .........................(((....(((((.......))))).))).. ( -8.10 = -8.10 + -0.00)



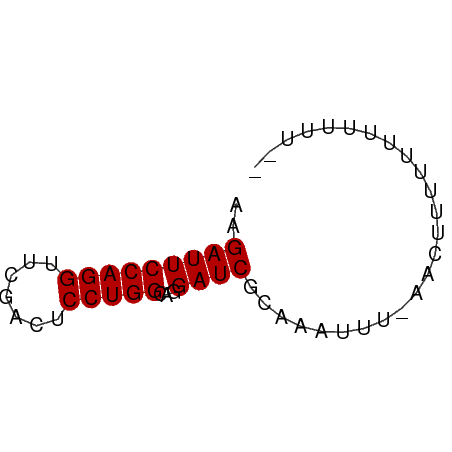
| Location | 2,066,068 – 2,066,119 |
|---|---|
| Length | 51 |
| Sequences | 7 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 78.82 |
| Shannon entropy | 0.43017 |
| G+C content | 0.39695 |
| Mean single sequence MFE | -10.89 |
| Consensus MFE | -9.40 |
| Energy contribution | -9.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2066068 51 - 21146708 AAGAUUCCAGGUUCGACUCCUGGCAGGAUCGUAUUC--GAAUUUUUUUAUUUU-- ..(((((((((.......)))))...))))......--...............-- ( -9.40, z-score = -0.55, R) >droYak2.chr2L 1912626 53 + 22324452 AAGAUUCCAGGUUCGACUCCUGGCAGGAUCGAAUUUUUACACUUUUUUGUUUU-- ........((((((((.(((.....))))))))))).................-- ( -9.60, z-score = -0.69, R) >droEre2.scaffold_4929 6374984 51 + 26641161 AAGAUUCCAGGUUCGACUCCUGGCAGGAUCGGACUUU--CACUUUUUUGUUUU-- ........((((((((.(((.....))))))))))).--..............-- ( -11.50, z-score = -0.73, R) >droPer1.super_73 202093 50 + 267513 AAGAUUCCAGGUUCGACUCCUGGCAGGAUCGGAGAU---AUCUUUUUUAUUUG-- ..(((((((((.......)))))...))))((((..---..))))........-- ( -9.70, z-score = 0.10, R) >droWil1.scaffold_180700 756544 54 + 6630534 AAGAUUCCAGGUUCGACUCCUGGCAGGAUCGCAGAUGUAUAAUUUUU-UUACAUU ..(((((((((.......)))))...))))...((((((........-.)))))) ( -11.90, z-score = -1.61, R) >droVir3.scaffold_12875 4798489 52 - 20611582 AAGAUUCCAGGUUCGACUCCUGGCAGGAUCGUUAAUUUUUUCUUUUCAUUAC--- ..(((((((((.......)))))...))))......................--- ( -9.40, z-score = -1.04, R) >droMoj3.scaffold_6496 1594950 52 + 26866924 AAGAUUCCAGGUUCGACUCCUGGCAGGAUCGCAAGAUUCAACUUUUUAUUAU--- ......(((((.......)))))..(((((....))))).............--- ( -14.70, z-score = -2.88, R) >consensus AAGAUUCCAGGUUCGACUCCUGGCAGGAUCGCAAAUUU_AACUUUUUUUUUUU__ ..(((((((((.......)))))...))))......................... ( -9.40 = -9.40 + 0.00)

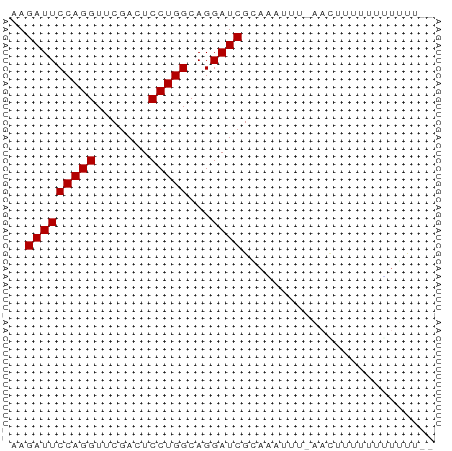
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:16 2011