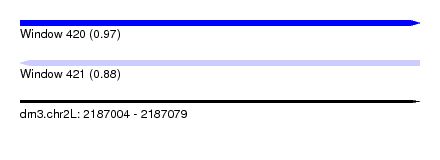
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,187,004 – 2,187,079 |
| Length | 75 |
| Max. P | 0.969231 |

| Location | 2,187,004 – 2,187,079 |
|---|---|
| Length | 75 |
| Sequences | 13 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 89.65 |
| Shannon entropy | 0.22094 |
| G+C content | 0.56039 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -24.89 |
| Energy contribution | -24.40 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
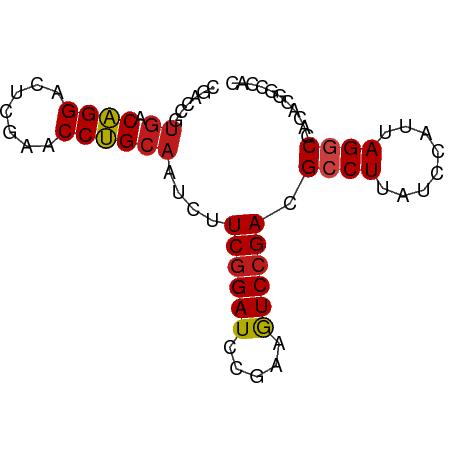
>dm3.chr2L 2187004 75 + 23011544 GUGGCCGUGUAGCCUAAUGGAUAAGGCGUCGGAUUUCUGAUCCGAAAAUUGCGGGUUCAAGUCCCGUCAUGGUCG ..(((((((..((((........)))).(((((((...))))))).....(((((.......)))))))))))). ( -29.30, z-score = -3.22, R) >droVir3.scaffold_12855 9965847 75 + 10161210 GGGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG ..(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))). ( -31.50, z-score = -2.64, R) >droWil1.scaffold_180949 1082149 75 + 6375548 AUGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAAUCCUGUCACGGUCG ..(((((((..((((........)))).((((((....).))))).....(((((.......)))))))))))). ( -31.40, z-score = -2.95, R) >dp4.chr2 7709197 75 - 30794189 GCGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG .((((((((..((((........)))).((((((....).))))).....(((((.......))))))))))))) ( -32.30, z-score = -2.63, R) >droAna3.scaffold_12916 9898646 74 - 16180835 -UGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG -.(((((((..((((........)))).((((((....).))))).....(((((.......)))))))))))). ( -31.40, z-score = -2.80, R) >droEre2.scaffold_4929 2223882 75 + 26641161 GUGGCUGUGUAGCCUAAUGGAUAAGGCGUCGGAUUUCUGAUCCGAAGAUUGCGGGUUCGAGUCCCGUCAUGGUCG ..(((((((..((((........)))).(((((((...))))))).....(((((.......)))))))))))). ( -26.40, z-score = -1.74, R) >droYak2.chr2L 2161986 75 + 22324452 GUGGCUGUGUAGCCUAAUGGAUAAGGCGUCGGAUUUCUGAUCCGAAGAUUGCGGGUUCGAGUCCCGUCAUGGUCG ..(((((((..((((........)))).(((((((...))))))).....(((((.......)))))))))))). ( -26.40, z-score = -1.74, R) >droSec1.super_121 59527 75 + 59987 GUGGCAGCGUAGGCUAAUGGAUAAGGCGUCGGAUUUCUGAUCCGAAAAUUGCGGGUUCAAGUCCCGUUAUGGUCG .((((.......))))........(((.(((((((...)))))))..((.(((((.......))))).)).))). ( -19.70, z-score = 0.05, R) >droSim1.chr2L 2140584 75 + 22036055 GUGGCCGUGUAGGCUAAUGGAUAAGGCGUCUGACUUCUGAUCCGAAAAUUGCGGGUUCAAGUCCCGUCCUGGUCA .(((((.....)))))...(((.(((((...(((((..((((((.......)))))).))))).)).))).))). ( -24.60, z-score = -1.51, R) >anoGam1.chr2R 51860471 73 + 62725911 --GGCCGUGUGGCCUAAUGGAGAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG --(((((((..((((........)))).((((((....).))))).....(((((.......)))))))))))). ( -30.80, z-score = -2.17, R) >droPer1.super_21 341708 71 - 1645244 ----CCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG ----(((((..((((........)))).((((((....).))))).....(((((.......))))))))))... ( -25.70, z-score = -1.34, R) >droMoj3.scaffold_6482 1037766 73 - 2735782 --GACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG --(((((((..((((........)))).((((((....).))))).....(((((.......)))))))))))). ( -31.40, z-score = -2.84, R) >droGri2.scaffold_15110 4377003 73 + 24565398 --GACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG --(((((((..((((........)))).((((((....).))))).....(((((.......)))))))))))). ( -31.40, z-score = -2.84, R) >consensus GUGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG ..(((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))). (-24.89 = -24.40 + -0.49)

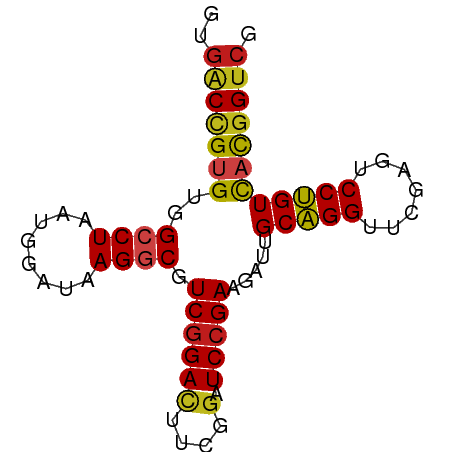
| Location | 2,187,004 – 2,187,079 |
|---|---|
| Length | 75 |
| Sequences | 13 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 89.65 |
| Shannon entropy | 0.22094 |
| G+C content | 0.56039 |
| Mean single sequence MFE | -23.41 |
| Consensus MFE | -15.57 |
| Energy contribution | -15.27 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2187004 75 - 23011544 CGACCAUGACGGGACUUGAACCCGCAAUUUUCGGAUCAGAAAUCCGACGCCUUAUCCAUUAGGCUACACGGCCAC .(.((.((.((((.......))))))....((((((.....)))))).((((........)))).....)).).. ( -20.30, z-score = -2.23, R) >droVir3.scaffold_12855 9965847 75 - 10161210 CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCCC .(((((((...(((((((((.(((.......))).)))).)))))...((((........))))..))))))).. ( -31.00, z-score = -4.53, R) >droWil1.scaffold_180949 1082149 75 - 6375548 CGACCGUGACAGGAUUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCAU .(((((((...(((((((((.........)))))))))..........((((........))))..))))))).. ( -29.70, z-score = -4.13, R) >dp4.chr2 7709197 75 + 30794189 CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCGC ((((((((.((((.......))))......((((((.....)))))).((((........))))..)))))))). ( -29.40, z-score = -3.56, R) >droAna3.scaffold_12916 9898646 74 + 16180835 CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCA- .(((((((.((((.......))))......((((((.....)))))).((((........))))..))))))).- ( -28.60, z-score = -3.67, R) >droEre2.scaffold_4929 2223882 75 - 26641161 CGACCAUGACGGGACUCGAACCCGCAAUCUUCGGAUCAGAAAUCCGACGCCUUAUCCAUUAGGCUACACAGCCAC ......((.((((.......))))))....((((((.....)))))).((((........))))........... ( -18.40, z-score = -1.87, R) >droYak2.chr2L 2161986 75 - 22324452 CGACCAUGACGGGACUCGAACCCGCAAUCUUCGGAUCAGAAAUCCGACGCCUUAUCCAUUAGGCUACACAGCCAC ......((.((((.......))))))....((((((.....)))))).((((........))))........... ( -18.40, z-score = -1.87, R) >droSec1.super_121 59527 75 - 59987 CGACCAUAACGGGACUUGAACCCGCAAUUUUCGGAUCAGAAAUCCGACGCCUUAUCCAUUAGCCUACGCUGCCAC .........((((.......))))......((((((.....))))))............((((....)))).... ( -13.50, z-score = -0.54, R) >droSim1.chr2L 2140584 75 - 22036055 UGACCAGGACGGGACUUGAACCCGCAAUUUUCGGAUCAGAAGUCAGACGCCUUAUCCAUUAGCCUACACGGCCAC .((..(((.((.(((((((..(((.......))).)))..))))...)))))..)).....(((.....)))... ( -15.80, z-score = -0.27, R) >anoGam1.chr2R 51860471 73 - 62725911 CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUCUCCAUUAGGCCACACGGCC-- ...(((((.((((.......))))......((((((.....)))))).((((........))))..)))))..-- ( -22.90, z-score = -1.70, R) >droPer1.super_21 341708 71 + 1645244 CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGG---- ...(((((.((((.......))))......((((((.....)))))).((((........))))..)))))---- ( -22.50, z-score = -2.23, R) >droMoj3.scaffold_6482 1037766 73 + 2735782 CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUC-- .(((((((.((((.......))))......((((((.....)))))).((((........))))..)))))))-- ( -26.90, z-score = -3.10, R) >droGri2.scaffold_15110 4377003 73 - 24565398 CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUC-- .(((((((.((((.......))))......((((((.....)))))).((((........))))..)))))))-- ( -26.90, z-score = -3.10, R) >consensus CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGCCAC ......((.((((.......))))))....((((((.....)))))).((((........))))........... (-15.57 = -15.27 + -0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:30 2011