
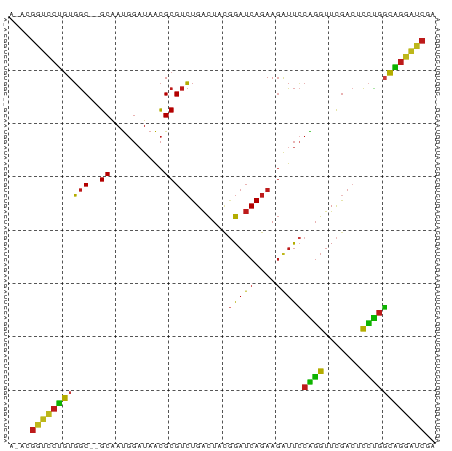
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,059,187 – 2,059,265 |
| Length | 78 |
| Max. P | 0.807896 |

| Location | 2,059,187 – 2,059,265 |
|---|---|
| Length | 78 |
| Sequences | 14 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 84.96 |
| Shannon entropy | 0.37086 |
| G+C content | 0.53543 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -19.88 |
| Energy contribution | -21.27 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
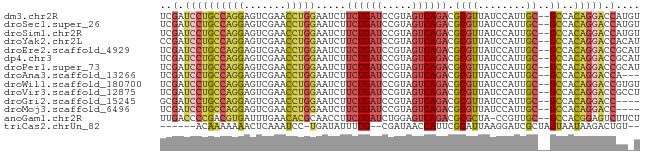
>dm3.chr2R 2059187 78 + 21146708 ACAUGGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA ...((((((((((((--((..........))))).......(((((....)))))((((.......))))))))))))). ( -26.90, z-score = -1.32, R) >droSec1.super_26 701335 78 + 826190 ACAUGGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA ...((((((((((((--((..........))))).......(((((....)))))((((.......))))))))))))). ( -26.90, z-score = -1.32, R) >droSim1.chr2R 953932 78 + 19596830 ACAUGGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA ...((((((((((((--((..........))))).......(((((....)))))((((.......))))))))))))). ( -26.90, z-score = -1.32, R) >droYak2.chr2L 1906229 78 - 22324452 AUGUGGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGG ...((((((((((((--((..........))))).......(((((....)))))((((.......))))))))))))). ( -26.80, z-score = -1.10, R) >droEre2.scaffold_4929 6368344 78 - 26641161 AUGCGGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA ...((((((((((((--((..........))))).......(((((....)))))((((.......))))))))))))). ( -28.60, z-score = -1.55, R) >dp4.chr3 17983685 78 + 19779522 AUGCGGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA ...((((((((((((--((..........))))).......(((((....)))))((((.......))))))))))))). ( -28.60, z-score = -1.55, R) >droPer1.super_73 194627 78 - 267513 AUGCGGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA ...((((((((((((--((..........))))).......(((((....)))))((((.......))))))))))))). ( -28.60, z-score = -1.55, R) >droAna3.scaffold_13266 1140944 75 - 19884421 ---UGGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA ---.(((((((((((--((..........))))).......(((((....)))))((((.......)))))))))))).. ( -26.60, z-score = -1.40, R) >droWil1.scaffold_180700 732170 78 - 6630534 ACACGGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA ...((((((((((((--((..........))))).......(((((....)))))((((.......))))))))))))). ( -28.90, z-score = -1.90, R) >droVir3.scaffold_12875 4787536 78 + 20611582 AGGCGGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA ...((((((((((((--((..........))))).......(((((....)))))((((.......))))))))))))). ( -28.60, z-score = -1.32, R) >droGri2.scaffold_15245 18126394 74 - 18325388 ----GGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGC ----(((((((((((--((..........))))).......(((((....)))))((((.......)))))))))))).. ( -26.60, z-score = -1.52, R) >droMoj3.scaffold_6496 1583425 74 - 26866924 ----GGUCCUGUGGC--GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA ----(((((((((((--((..........))))).......(((((....)))))((((.......)))))))))))).. ( -26.60, z-score = -1.44, R) >anoGam1.chr2R 5380044 77 - 62725911 AGAAGACUCCGUGGC--GCAACGG-UAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAA ....(((((((..((--((.....-..)))).((((((....).))))).....(((((.......)))))))))))).. ( -26.60, z-score = -1.35, R) >triCas2.chrUn_82 83276 69 + 117267 --ACAGUCUUAUUACUAGCGAUCCUUAAUGCGAAUGGUUAUCG--CAAAAUAUCA-GGAUUUGAGUUUUUUUGU------ --((((....(..(((...((((((...(((((.......)))--)).......)-)))))..)))..).))))------ ( -11.00, z-score = -0.27, R) >consensus A_ACGGUCCUGUGGC__GCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA ....(((((((.....................((((((....).))))).....(((((.......)))))))))))).. (-19.88 = -21.27 + 1.39)


| Location | 2,059,187 – 2,059,265 |
|---|---|
| Length | 78 |
| Sequences | 14 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 84.96 |
| Shannon entropy | 0.37086 |
| G+C content | 0.53543 |
| Mean single sequence MFE | -23.02 |
| Consensus MFE | -16.06 |
| Energy contribution | -16.19 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2059187 78 - 21146708 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACCAUGU ..(.((((((((((.......))))).....((((((.....)))))).((((........))--))..))))).).... ( -23.80, z-score = -1.40, R) >droSec1.super_26 701335 78 - 826190 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACCAUGU ..(.((((((((((.......))))).....((((((.....)))))).((((........))--))..))))).).... ( -23.80, z-score = -1.40, R) >droSim1.chr2R 953932 78 - 19596830 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACCAUGU ..(.((((((((((.......))))).....((((((.....)))))).((((........))--))..))))).).... ( -23.80, z-score = -1.40, R) >droYak2.chr2L 1906229 78 + 22324452 CCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACCACAU ..(.((((((((((.......))))).....((((((.....)))))).((((........))--))..))))).).... ( -23.80, z-score = -1.82, R) >droEre2.scaffold_4929 6368344 78 + 26641161 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACCGCAU .((.((((((((((.......))))).....((((((.....)))))).((((........))--))..))))).))... ( -24.70, z-score = -1.64, R) >dp4.chr3 17983685 78 - 19779522 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACCGCAU .((.((((((((((.......))))).....((((((.....)))))).((((........))--))..))))).))... ( -24.70, z-score = -1.64, R) >droPer1.super_73 194627 78 + 267513 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACCGCAU .((.((((((((((.......))))).....((((((.....)))))).((((........))--))..))))).))... ( -24.70, z-score = -1.64, R) >droAna3.scaffold_13266 1140944 75 + 19884421 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACCA--- ..(.((((((((((.......))))).....((((((.....)))))).((((........))--))..))))).).--- ( -23.80, z-score = -1.75, R) >droWil1.scaffold_180700 732170 78 + 6630534 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACCGUGU .((.((((((((((.......))))).....((((((.....)))))).((((........))--))..))))).))... ( -24.90, z-score = -1.46, R) >droVir3.scaffold_12875 4787536 78 - 20611582 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACCGCCU .((.((((((((((.......))))).....((((((.....)))))).((((........))--))..))))).))... ( -24.70, z-score = -1.57, R) >droGri2.scaffold_15245 18126394 74 + 18325388 GCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACC---- ....((((((((((.......))))).....((((((.....)))))).((((........))--))..)))))..---- ( -23.20, z-score = -1.45, R) >droMoj3.scaffold_6496 1583425 74 + 26866924 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC--GCCACAGGACC---- ....((((((((((.......))))).....((((((.....)))))).((((........))--))..)))))..---- ( -23.20, z-score = -1.60, R) >anoGam1.chr2R 5380044 77 + 62725911 UUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUA-CCGUUGC--GCCACGGAGUCUUCU ..(((.(((.((((.......))))......((((((.....)))))).((((..-.....))--))..))).))).... ( -22.20, z-score = -1.29, R) >triCas2.chrUn_82 83276 69 - 117267 ------ACAAAAAAACUCAAAUCC-UGAUAUUUUG--CGAUAACCAUUCGCAUUAAGGAUCGCUAGUAAUAAGACUGU-- ------........(((...((((-(.......((--(((.......)))))...)))))....)))...........-- ( -11.00, z-score = -1.64, R) >consensus UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGC__GCCACAGGACCGU_U ..(.((((((((((.......))))).....((((((.....)))))).....................))))).).... (-16.06 = -16.19 + 0.12)
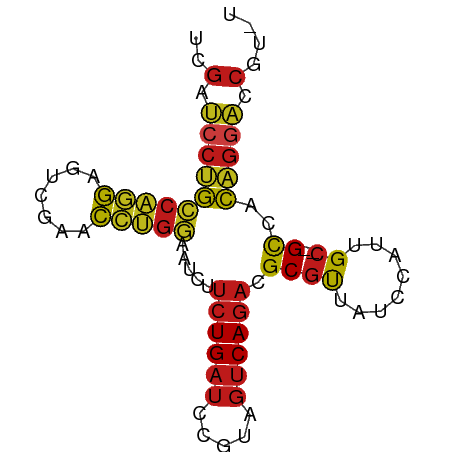

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:13 2011