
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,059,000 – 2,059,077 |
| Length | 77 |
| Max. P | 0.997740 |

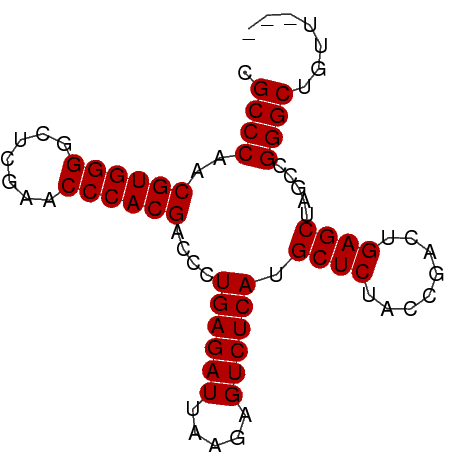
| Location | 2,059,000 – 2,059,077 |
|---|---|
| Length | 77 |
| Sequences | 11 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Shannon entropy | 0.08424 |
| G+C content | 0.60486 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -31.07 |
| Energy contribution | -31.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.14 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.997740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
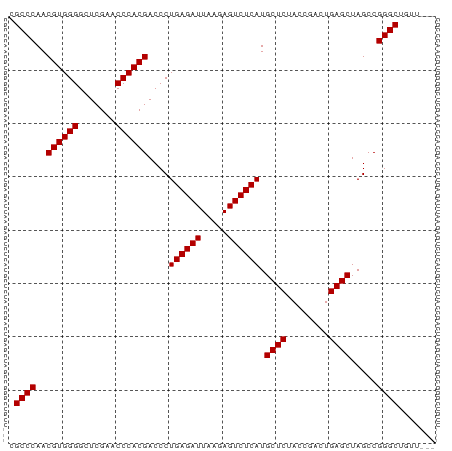
>dm3.chr2R 2059000 77 + 21146708 CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUU--- .((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....--- ( -31.00, z-score = -3.05, R) >droVir3.scaffold_12875 15813170 80 - 20611582 CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUUUGGC .((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....... ( -31.50, z-score = -2.80, R) >droGri2.scaffold_15245 189453 77 + 18325388 CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGUU--- .((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....--- ( -31.50, z-score = -3.14, R) >droMoj3.scaffold_6496 25272272 77 + 26866924 CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUU--- .((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....--- ( -31.00, z-score = -3.05, R) >droSec1.super_26 701148 77 + 826190 CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGCU--- .((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....--- ( -31.00, z-score = -2.99, R) >droSim1.chr2R 953745 77 + 19596830 CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUU--- .((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....--- ( -31.00, z-score = -3.05, R) >droYak2.chr2L 1906042 77 - 22324452 CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUC--- .((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....--- ( -31.00, z-score = -2.87, R) >droEre2.scaffold_4929 6368157 77 - 26641161 CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUG--- .((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....--- ( -31.00, z-score = -3.09, R) >droAna3.scaffold_13266 18747157 73 + 19884421 CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC------- .((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))------- ( -29.80, z-score = -3.21, R) >dp4.chr3 17983382 74 + 19779522 CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCA------ .((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).------ ( -31.50, z-score = -3.64, R) >droPer1.super_73 194315 74 - 267513 CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCA------ .((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).------ ( -31.50, z-score = -3.64, R) >consensus CGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUU___ .((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....... (-31.07 = -31.07 + 0.00)

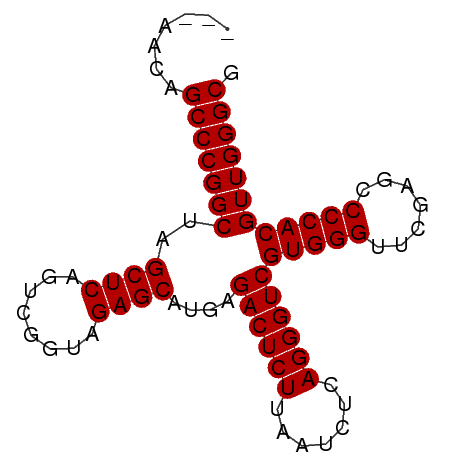
| Location | 2,059,000 – 2,059,077 |
|---|---|
| Length | 77 |
| Sequences | 11 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Shannon entropy | 0.08424 |
| G+C content | 0.60486 |
| Mean single sequence MFE | -32.81 |
| Consensus MFE | -32.81 |
| Energy contribution | -32.81 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
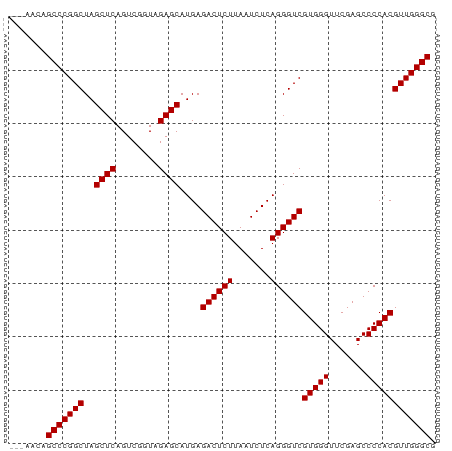
>dm3.chr2R 2059000 77 - 21146708 ---AACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG ---....(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). ( -32.90, z-score = -2.62, R) >droVir3.scaffold_12875 15813170 80 + 20611582 GCCAAAUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG .......(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). ( -32.70, z-score = -2.15, R) >droGri2.scaffold_15245 189453 77 - 18325388 ---AACUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG ---....(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). ( -32.70, z-score = -2.29, R) >droMoj3.scaffold_6496 25272272 77 - 26866924 ---AACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG ---....(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). ( -32.90, z-score = -2.62, R) >droSec1.super_26 701148 77 - 826190 ---AGCAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG ---....(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). ( -32.90, z-score = -2.25, R) >droSim1.chr2R 953745 77 - 19596830 ---AACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG ---....(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). ( -32.90, z-score = -2.62, R) >droYak2.chr2L 1906042 77 + 22324452 ---GACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG ---....(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). ( -32.90, z-score = -2.29, R) >droEre2.scaffold_4929 6368157 77 + 26641161 ---CACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG ---....(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). ( -32.90, z-score = -2.58, R) >droAna3.scaffold_13266 18747157 73 - 19884421 -------GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG -------(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). ( -32.70, z-score = -2.71, R) >dp4.chr3 17983382 74 - 19779522 ------UGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG ------.(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). ( -32.70, z-score = -2.55, R) >droPer1.super_73 194315 74 + 267513 ------UGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG ------.(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). ( -32.70, z-score = -2.55, R) >consensus ___AACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCG .......(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))). (-32.81 = -32.81 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:11 2011