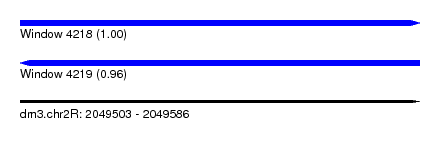
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,049,503 – 2,049,586 |
| Length | 83 |
| Max. P | 0.998493 |

| Location | 2,049,503 – 2,049,586 |
|---|---|
| Length | 83 |
| Sequences | 12 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 58.21 |
| Shannon entropy | 0.88656 |
| G+C content | 0.47129 |
| Mean single sequence MFE | -19.68 |
| Consensus MFE | -10.75 |
| Energy contribution | -9.92 |
| Covariance contribution | -0.82 |
| Combinations/Pair | 2.64 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.998493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
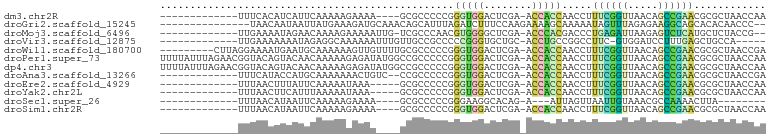
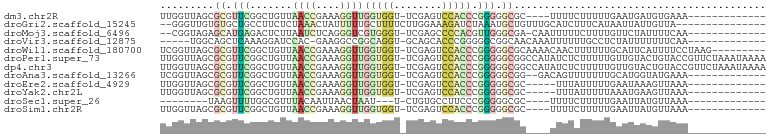
>dm3.chr2R 2049503 83 + 21146708 -------------UUUCACAUCAUUCAAAAAGAAAA----GCGCCCCCGGGUGGACUCGA-ACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAA -------------......................(----((((.....(((((......-.))))).....((((((.....)))))).)))))...... ( -22.80, z-score = -2.94, R) >droGri2.scaffold_15245 18116282 84 - 18325388 ---------------UAACAAUAAUUAUGAAAGAUGCAAACAGCAUUUAGAUCUUUCCAAGAAAAGCAAAAAUAGUUUAGAGAAGGCAGCACACAACCC-- ---------------................((((((.....)))))).(..(((((......((((.......))))...)))))...).........-- ( -6.60, z-score = 0.23, R) >droMoj3.scaffold_6496 25272245 84 + 26866924 -------------UUGAAAAUAGAACAAAAGAAAAAUUG-UCGCCCAACGUGGGGCUCGA-ACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCG-- -------------.......((((.((............-........((((((......-.))))))....((((((.....)))))))).))))...-- ( -20.10, z-score = -2.23, R) >droVir3.scaffold_12875 15788720 81 - 20611582 -------------UUGAAAAAAAUAGAGGCAAAAAAUUUGUUGCCGCCCCCGGGUGCUGC-ACCUGCCGGCCUUC-GUGGAUCCUUUGAGCUGCCA----- -------------..(((.........(((((........)))))(((..((((((...)-)))))..))).)))-(..(.((....)).)..)..----- ( -21.00, z-score = 0.36, R) >droWil1.scaffold_180700 720713 91 - 6630534 ---------CUUAGGAAAAUGAAUGCAAAAAAGUUGUUUUGCGCCCCCGGGUGGACUCGA-ACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGA ---------...(((.........((((((......)))))).......(((((......-.)))))..))).(((((((...((.....))..))))))) ( -23.80, z-score = -0.68, R) >droPer1.super_73 183836 100 - 267513 UUUUAUUUAGAACGGUACAGUACAACAAAAAGAGAUAUGGCCGCCCCCGGGUGGACUCGA-ACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAA .............(((..((..(........(((......(((((....))))).)))..-...........((((((.....)))))).)..)).))).. ( -22.50, z-score = -0.36, R) >dp4.chr3 17972884 100 + 19779522 UUUUAUUUAGAACGGUACAGUACAACAAAAAGAGAUAUGGCCGCCCCCGGGUGGACUCGA-ACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAA .............(((..((..(........(((......(((((....))))).)))..-...........((((((.....)))))).)..)).))).. ( -22.50, z-score = -0.36, R) >droAna3.scaffold_13266 1134695 85 - 19884421 -------------UUUCAUACCAUGCAAAAAAACUGUC--CCGCCCCCGGGUGGACUCGA-ACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGA -------------...........((.........(((--(.(((....)))))))....-...........((((((.....)))))).))......... ( -19.00, z-score = -0.76, R) >droEre2.scaffold_4929 6358825 82 - 26641161 -------------UUUAACUUUAUUCAAAAAUAAA-----GCGCCCCCGGGUGGACUCGA-ACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAA -------------.....................(-----((((.....(((((......-.))))).....((((((.....)))))).)))))...... ( -22.80, z-score = -3.25, R) >droYak2.chr2L 1895776 82 - 22324452 -------------UUUAACUUCAUUUAAAAAUAAA-----GCGCCCCCGGGUGGACUCGA-ACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAA -------------.....................(-----((((.....(((((......-.))))).....((((((.....)))))).)))))...... ( -22.80, z-score = -3.25, R) >droSec1.super_26 691928 72 + 826190 -------------UUUAACAUAAUUCAAAAAGAAAA----GCGCCCCCGGGAAGGCACAG-A---AUUAGUUAAUUGUAAACGCCAAAACUUA-------- -------------.(((((.((((((.....(....----.)(((........)))...)-)---)))))))))...................-------- ( -9.50, z-score = -0.11, R) >droSim1.chr2R 944679 83 + 19596830 -------------UUUAACAUAAUUCAAAAAGAAAA----GCGCCCCCGGGUGGACUCGA-ACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAA -------------......................(----((((.....(((((......-.))))).....((((((.....)))))).)))))...... ( -22.80, z-score = -3.12, R) >consensus _____________UUUAACAUAAUUCAAAAAAAAAA____GCGCCCCCGGGUGGACUCGA_ACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAA .................................................(((((........))))).....((((((.....))))))............ (-10.75 = -9.92 + -0.82)
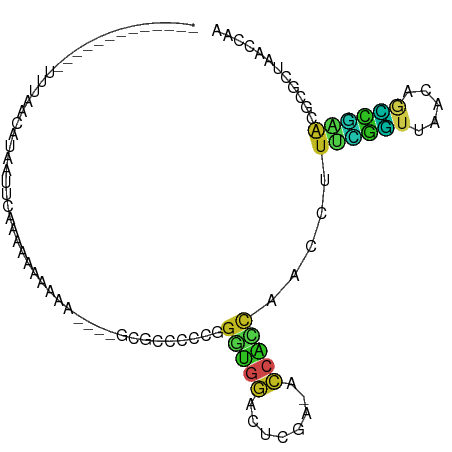
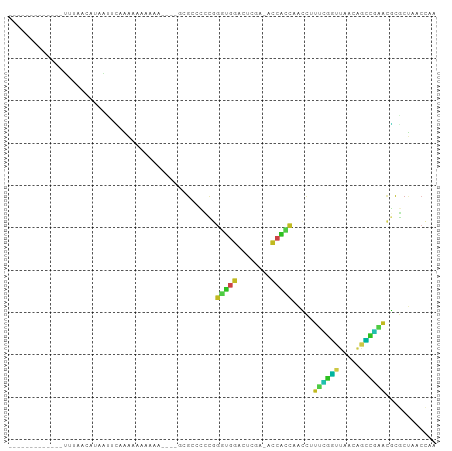
| Location | 2,049,503 – 2,049,586 |
|---|---|
| Length | 83 |
| Sequences | 12 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 58.21 |
| Shannon entropy | 0.88656 |
| G+C content | 0.47129 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.75 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 2.14 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
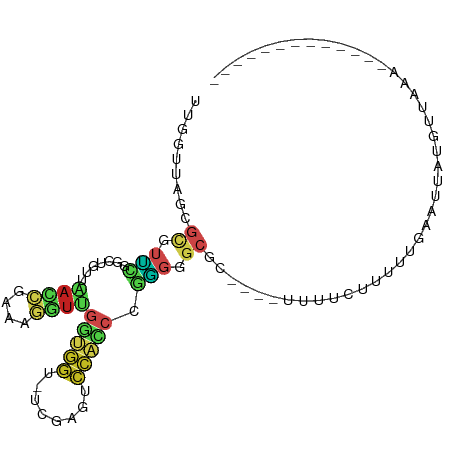
>dm3.chr2R 2049503 83 - 21146708 UUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGU-UCGAGUCCACCCGGGGGCGC----UUUUCUUUUUGAAUGAUGUGAAA------------- ..((..(((((.(((((....(((((....)))))((((.-......))))))))).))))----)..))..................------------- ( -29.50, z-score = -1.94, R) >droGri2.scaffold_15245 18116282 84 + 18325388 --GGGUUGUGUGCUGCCUUCUCUAAACUAUUUUUGCUUUUCUUGGAAAGAUCUAAAUGCUGUUUGCAUCUUUCAUAAUUAUUGUUA--------------- --((((.(....).))))..........................(((((((.(((((...))))).))))))).............--------------- ( -10.80, z-score = 0.21, R) >droMoj3.scaffold_6496 25272245 84 - 26866924 --CGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGU-UCGAGCCCCACGUUGGGCGA-CAAUUUUUCUUUUGUUCUAUUUUCAA------------- --.(((((((((.((((((((.......))))))(((((.-......)))))(((....))-)......))...))))))))).....------------- ( -24.20, z-score = -1.58, R) >droVir3.scaffold_12875 15788720 81 + 20611582 -----UGGCAGCUCAAAGGAUCCAC-GAAGGCCGGCAGGU-GCAGCACCCGGGGGCGGCAACAAAUUUUUUGCCUCUAUUUUUUUCAA------------- -----.((((((((.......((..-...))((((...((-(...)))))))))))(....)........))))..............------------- ( -18.80, z-score = 1.65, R) >droWil1.scaffold_180700 720713 91 + 6630534 UCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGU-UCGAGUCCACCCGGGGGCGCAAAACAACUUUUUUGCAUUCAUUUUCCUAAG--------- (((((((((((.....)).))))))))).(((..(((((.-....((((......))))((((((......))))))..)))))..)))...--------- ( -30.90, z-score = -2.04, R) >droPer1.super_73 183836 100 + 267513 UUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGU-UCGAGUCCACCCGGGGGCGGCCAUAUCUCUUUUUGUUGUACUGUACCGUUCUAAAUAAAA ..((((((..((...(((((((((((....))))(((((.-......)))))....)))))))(((........)))))..))).)))............. ( -29.50, z-score = -0.50, R) >dp4.chr3 17972884 100 - 19779522 UUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGU-UCGAGUCCACCCGGGGGCGGCCAUAUCUCUUUUUGUUGUACUGUACCGUUCUAAAUAAAA ..((((((..((...(((((((((((....))))(((((.-......)))))....)))))))(((........)))))..))).)))............. ( -29.50, z-score = -0.50, R) >droAna3.scaffold_13266 1134695 85 + 19884421 UCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGU-UCGAGUCCACCCGGGGGCGG--GACAGUUUUUUUGCAUGGUAUGAAA------------- .........(((.(((((..((((((....))))))..).-))))((((.((....))..)--)))........)))...........------------- ( -27.40, z-score = -0.56, R) >droEre2.scaffold_4929 6358825 82 + 26641161 UUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGU-UCGAGUCCACCCGGGGGCGC-----UUUAUUUUUGAAUAAAGUUAAA------------- ......(((((.(((((....(((((....)))))((((.-......))))))))).))))-----).....................------------- ( -27.40, z-score = -1.86, R) >droYak2.chr2L 1895776 82 + 22324452 UUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGU-UCGAGUCCACCCGGGGGCGC-----UUUAUUUUUAAAUGAAGUUAAA------------- ......(((((.(((((....(((((....)))))((((.-......))))))))).))))-----).....................------------- ( -27.40, z-score = -1.86, R) >droSec1.super_26 691928 72 - 826190 --------UAAGUUUUGGCGUUUACAAUUAACUAAU---U-CUGUGCCUUCCCGGGGGCGC----UUUUCUUUUUGAAUUAUGUUAAA------------- --------...................(((((((((---(-(.(((((((....)))))))----..........)))))).))))).------------- ( -15.90, z-score = -1.42, R) >droSim1.chr2R 944679 83 - 19596830 UUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGU-UCGAGUCCACCCGGGGGCGC----UUUUCUUUUUGAAUUAUGUUAAA------------- ..((..(((((.(((((....(((((....)))))((((.-......))))))))).))))----)..))..................------------- ( -29.50, z-score = -2.36, R) >consensus UUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGU_UCGAGUCCACCCGGGGGCGC____UUUUCUUUUUGAAUUAUGUUAAA_____________ .........((.(((.......((((....))))(((((........))))).))).)).......................................... (-10.55 = -10.75 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:10 2011