| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,049,038 – 2,049,122 |
| Length | 84 |
| Max. P | 0.987345 |

| Location | 2,049,038 – 2,049,122 |
|---|---|
| Length | 84 |
| Sequences | 11 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 71.58 |
| Shannon entropy | 0.61482 |
| G+C content | 0.49994 |
| Mean single sequence MFE | -21.55 |
| Consensus MFE | -14.81 |
| Energy contribution | -14.55 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
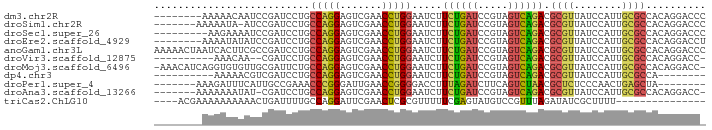
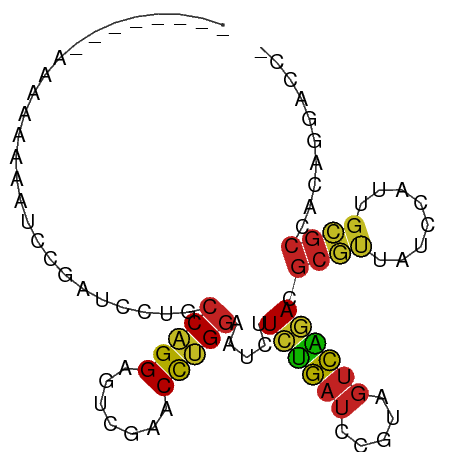
>dm3.chr2R 2049038 84 - 21146708 --------AAAAACAAUCCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCC --------.............((((((((((.......))))).....((((((.....)))))).((((........))))..)))))... ( -23.20, z-score = -1.89, R) >droSim1.chr2R 944169 84 - 19596830 -------AAAAAUA-AUCCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCC -------.......-......((((((((((.......))))).....((((((.....)))))).((((........))))..)))))... ( -23.20, z-score = -2.01, R) >droSec1.super_26 691413 83 - 826190 ---------AAGAAAAUCCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCC ---------............((((((((((.......))))).....((((((.....)))))).((((........))))..)))))... ( -23.20, z-score = -1.52, R) >droEre2.scaffold_4929 6358724 84 + 26641161 --------AAAAUAUAUCCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCU --------...........(.((((((((((.......))))).....((((((.....)))))).((((........))))..))))).). ( -23.30, z-score = -1.81, R) >anoGam1.chr3L 7649543 92 + 41284009 AAAAACUAAUCACUUCGCCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCC .....................((((((((((.......))))).....((((((.....)))))).((((........))))..)))))... ( -23.20, z-score = -1.28, R) >droVir3.scaffold_12875 4777441 79 - 20611582 ----------AAACAA--CGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACC- ----------......--...((((((((((.......))))).....((((((.....)))))).((((........))))..)))))..- ( -23.20, z-score = -1.88, R) >droMoj3.scaffold_6496 1573692 90 + 26866924 -AAACAUCAGGUGUGUUGCGAUUCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACC- -........(((......(((((((....)))))))..((((......((((((.....)))))).((((........))))..)))))))- ( -25.60, z-score = -0.54, R) >dp4.chr3 17972706 74 - 19779522 ----------AAAAACGUCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCA-------- ----------.........(((....(((((.......))))).))).((((((.....)))))).((((........))))..-------- ( -17.90, z-score = -0.62, R) >droPer1.super_4 1118579 77 + 7162766 -------AAAGAUUUCAUUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUA-------- -------.....((((......))))(((((.......))))).....((((((.....)))))).((((........))))..-------- ( -19.90, z-score = -1.12, R) >droAna3.scaffold_13266 18741865 83 - 19884421 -------AAAAAAAUAU-CGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACC- -------..........-...((((((((((.......))))).....((((((.....)))))).((((........))))..)))))..- ( -23.20, z-score = -2.08, R) >triCas2.ChLG10 5293633 73 - 8806720 ----ACGAAAAAAAAAACUGAUUUUGCCAGGAUUCGAACUCGCGUUUUUCGAGUAUGUCCGUUUAGAUAUCGCUUUU--------------- ----.........((((.((((((((...((((....(((((.......)))))..))))...)))).)))).))))--------------- ( -11.20, z-score = 0.81, R) >consensus ________AAAAAAAAUCCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACC_ ..........................(((((.......))))).....((((((.....)))))).((((........)))).......... (-14.81 = -14.55 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:08 2011