
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,040,972 – 2,041,084 |
| Length | 112 |
| Max. P | 0.991164 |

| Location | 2,040,972 – 2,041,084 |
|---|---|
| Length | 112 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.10 |
| Shannon entropy | 0.43585 |
| G+C content | 0.52056 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -29.09 |
| Energy contribution | -29.19 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990397 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
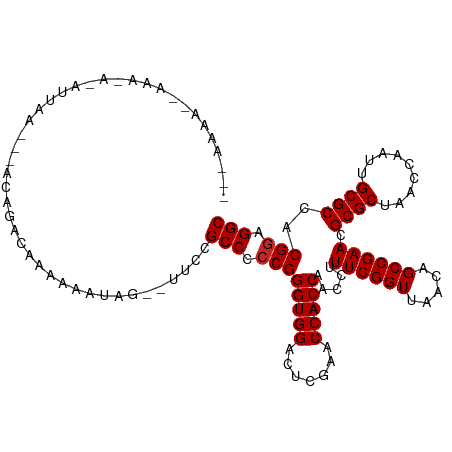
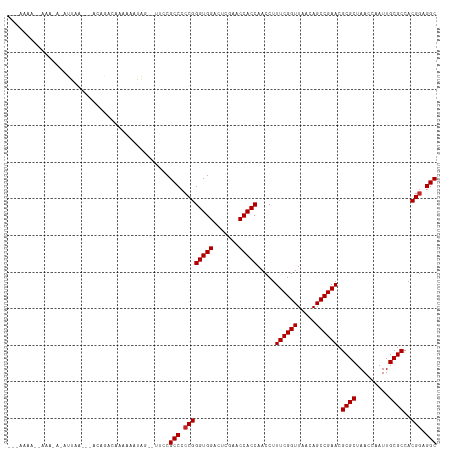
>dm3.chr2R 2040972 112 + 21146708 --AAAAAACAAAGAGAUUCC---ACAGGUAAAAUAGAAAUAUUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC --..................---........((((....)))).(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -30.30, z-score = -1.83, R) >droSim1.chr2R 936414 111 + 19596830 --AAAAAACAAAGAGAUUC----ACAGGUAAAAUAGAAAUAUUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC --.................----........((((....)))).(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -30.30, z-score = -1.92, R) >droSec1.super_26 683774 111 + 826190 --AAAAUACAAAGAGAUUC----ACAGGUAAAAUAGAAAUAUCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC --.................----...((....(((....)))))(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -30.60, z-score = -1.99, R) >droYak2.chr2L 1887093 103 - 22324452 ---------AGUAAAACUAA---CAAGAUAAAAAGAAG--UAUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC ---------...........---...((((........--))))(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -31.00, z-score = -2.48, R) >droEre2.scaffold_4929 6349456 117 - 26641161 GAAAAAAGUAUAUAUAUUAAAGGGCGGCCAAACUAAAAAAUGCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC .....................(((((((.............)))))))((((((((.......))))).....((((((.....)))))).((((.........))))..))).... ( -38.42, z-score = -2.51, R) >dp4.chr3 17964831 110 + 19779522 --GAAAAUUUGGAAAAGAGA---AUACAAAAAAAGUUG--UUCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC --........(((.......---..((((......)))--))))(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -31.30, z-score = -1.44, R) >droPer1.super_73 175823 110 - 267513 --GAAAAAUGAGAAAAGAGA---AUACAAAAAAGGUUG--UUCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC --......((((........---..((((......)))--)((((((....))))))))))..((.((.....((((((.....)))))).((((.........))))...)).)). ( -30.90, z-score = -1.36, R) >droWil1.scaffold_180700 708554 99 - 6630534 ---------------AAAAA---AUCAACAAAAAGUUGCUUUGCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGC ---------------...((---(.((((.....)))).)))..(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -31.90, z-score = -1.82, R) >droVir3.scaffold_12875 15844755 97 - 20611582 ---------------AAAAA---AAAAGCAAAAAGUUG--UUCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGC ---------------.....---...(((((....)))--))..(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -30.30, z-score = -1.44, R) >anoGam1.chr2L 39583674 93 - 48795086 -------------------A---ACAAACAAAAAACUU--CACCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGAAGGC -------------------.---............(((--(.(((((....))))).................((((((.....)))))).((((.........))))...)))).. ( -26.20, z-score = -1.99, R) >consensus ___AAAA__AAA_A_AUUAA___ACAGACAAAAAAUAG__UUCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC ............................................(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) (-29.09 = -29.19 + 0.10)

| Location | 2,040,972 – 2,041,084 |
|---|---|
| Length | 112 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.10 |
| Shannon entropy | 0.43585 |
| G+C content | 0.52056 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -36.71 |
| Energy contribution | -36.62 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991164 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
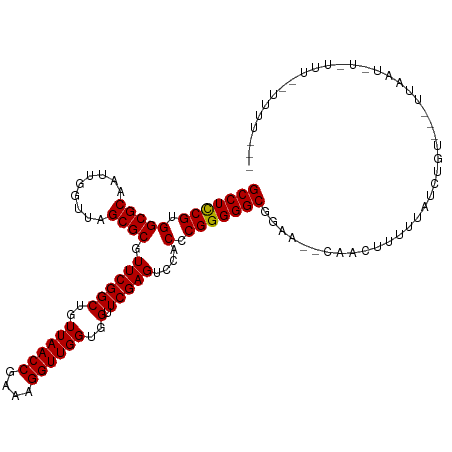
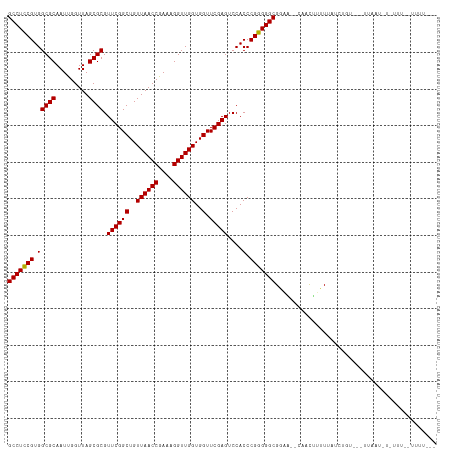
>dm3.chr2R 2040972 112 - 21146708 GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGAAUAUUUCUAUUUUACCUGU---GGAAUCUCUUUGUUUUUU-- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))......((((((.......))---))))..............-- ( -39.90, z-score = -2.28, R) >droSim1.chr2R 936414 111 - 19596830 GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGAAUAUUUCUAUUUUACCUGU----GAAUCUCUUUGUUUUUU-- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))(((((.......((((....)----)))......)))))...-- ( -37.22, z-score = -1.75, R) >droSec1.super_26 683774 111 - 826190 GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGAUAUUUCUAUUUUACCUGU----GAAUCUCUUUGUAUUUU-- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))((((((......((((....)----))).......)))))).-- ( -38.22, z-score = -1.76, R) >droYak2.chr2L 1887093 103 + 22324452 GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGAUA--CUUCUUUUUAUCUUG---UUAGUUUUACU--------- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))((((--........))))...---...........--------- ( -37.80, z-score = -2.32, R) >droEre2.scaffold_4929 6349456 117 + 26641161 GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGCAUUUUUUAGUUUGGCCGCCCUUUAAUAUAUAUACUUUUUUC (((.....)))......(((((((((.....)).)))))))(((((((.(((((.......))))).(((((((((.............)))))))))..........))))))).. ( -41.92, z-score = -1.76, R) >dp4.chr3 17964831 110 - 19779522 GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGAA--CAACUUUUUUUGUAU---UCUCUUUUCCAAAUUUUC-- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))((((--(((......))))..---.......)))........-- ( -38.80, z-score = -2.19, R) >droPer1.super_73 175823 110 + 267513 GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGAA--CAACCUUUUUUGUAU---UCUCUUUUCUCAUUUUUC-- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))((((--(((......)))).)---))................-- ( -38.30, z-score = -2.10, R) >droWil1.scaffold_180700 708554 99 + 6630534 GCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGCAAAGCAACUUUUUGUUGAU---UUUUU--------------- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))......((((.....))))..---.....--------------- ( -39.80, z-score = -2.20, R) >droVir3.scaffold_12875 15844755 97 + 20611582 GCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGAA--CAACUUUUUGCUUUU---UUUUU--------------- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))....--...............---.....--------------- ( -36.90, z-score = -1.52, R) >anoGam1.chr2L 39583674 93 + 48795086 GCCUUCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGUG--AAGUUUUUUGUUUGU---U------------------- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))....--...............---.------------------- ( -34.10, z-score = -1.25, R) >consensus GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGAA__CAACUUUUUAUCUGU___UUAAU_U_UUU__UUUU___ (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))............................................ (-36.71 = -36.62 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:06 2011