| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,040,089 – 2,040,202 |
| Length | 113 |
| Max. P | 0.997099 |

| Location | 2,040,089 – 2,040,202 |
|---|---|
| Length | 113 |
| Sequences | 12 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 75.39 |
| Shannon entropy | 0.55069 |
| G+C content | 0.53433 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.996934 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
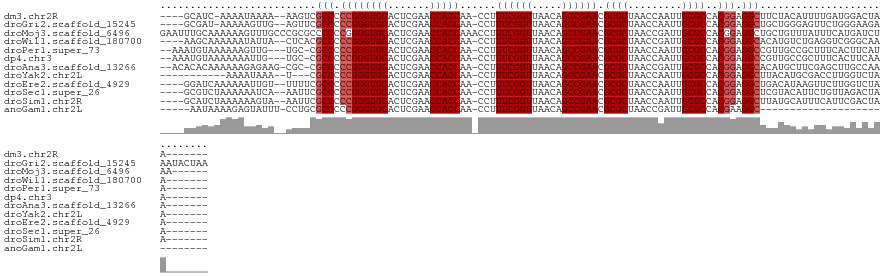
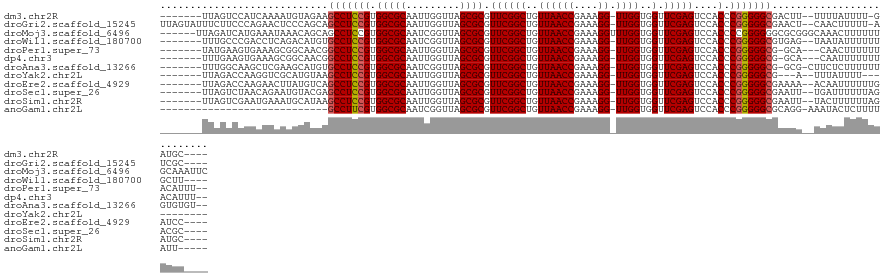
>dm3.chr2R 2040089 113 + 21146708 ----GCAUC-AAAAUAAAA--AAGUCGCCCCCGGGUGGACUCGAACCACCAA-CCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUUCUACAUUUUGAUGGACUAA------- ----.((((-(((((....--.((..(((.((((((((.......)))))..-...((((((.....)))))).((((.........))))..))).)))..))..)))))))))......------- ( -38.90, z-score = -4.10, R) >droGri2.scaffold_15245 208578 120 + 18325388 ----GCGAU-AAAAAGUUG--AGUUCGCCCCCGGGUGGACUCGAACCACCAA-CCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUGCUGGGAGUUCUGGGAAGAAAUACUAA ----.....-........(--((((((((....)))))))))((((..((((-((....))))..(((((....((((.........))))......)))))..))..))))................ ( -40.70, z-score = -1.45, R) >droMoj3.scaffold_6496 25253680 122 + 26866924 GAAUUUGCAAAAAAGUUUGCCCGCGCCCCCCGGGGUGGACUCGAACCACCAAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCUGCUGUUUAUUUCAUGAUCUAA------ (((...(((((....)))))..(((((..(((.(((((.......)))))......((((((.....)))))).((((.........))))..))).))).)).......))).........------ ( -34.90, z-score = -0.48, R) >droWil1.scaffold_180700 707023 114 - 6630534 ----AAGCAAAAAAUAUUA--CUCACGCCCCCGGGUGGACUCGAACCACCAA-CCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCACAUGUCUGAGGUCGGGCAAA------- ----...............--.....(((.((((((((.......)))))..-...((((((.....)))))).((((.........))))..))).)))...((((((....))))))..------- ( -38.00, z-score = -1.71, R) >droPer1.super_73 175288 114 - 267513 --AAAUGUAAAAAAGUUG---UGC-CGCCCCCGGGUGGACUCGAACCACCAA-CCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCGUUGCCGCUUUCACUUCAUA------- --..(((....(((((.(---(..-((((.((((((((.......)))))..-...((((((.....)))))).((((.........))))..))).)).))..)).))))).....))).------- ( -36.10, z-score = -1.56, R) >dp4.chr3 17964291 114 + 19779522 --AAAUGUAAAAAAAUUG---UGC-CGCCCCCGGGUGGACUCGAACCACCAA-CCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCGUUGCCGCUUUCACUUCAAA------- --................---.((-.(((.((((((((.......)))))..-...((((((.....)))))).((((.........))))..))).))).))..................------- ( -31.20, z-score = -0.39, R) >droAna3.scaffold_13266 1127335 116 - 19884421 --ACACACAAAAAAGAGAAG-CGC-CGCCCCCGGGUGGACUCGAACCACCAA-CCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCACAUGCUUCGAGCUUGCCAAA------- --..........(((.((((-((.-.(((.((((((((.......)))))..-...((((((.....)))))).((((.........))))..))).)))...))))))...)))......------- ( -39.50, z-score = -2.64, R) >droYak2.chr2L 1886453 104 - 22324452 -----------AAAAUAAA--U---CGCCCCCGGGUGGACUCGAACCACCAA-CCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUUACAUGCGACCUUGGUCUAA------- -----------........--.---.(((.((((((((.......)))))..-...((((((.....)))))).((((.........))))..))).)))........(((....)))...------- ( -34.20, z-score = -2.55, R) >droEre2.scaffold_4929 6348498 114 - 26641161 ----GGAUCAAAAAAUUGU--UUUUCGCCCCCGGGUGGACUCGAACCACCAA-CCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUGACAUAAGUUCUUGGUCUAA------- ----(((((((.((..(((--(....(((.((((((((.......)))))..-...((((((.....)))))).((((.........))))..))).))).))))....)).)))))))..------- ( -38.80, z-score = -2.80, R) >droSec1.super_26 682912 114 + 826190 ----GCGUCUAAAAAAUCA--AAUUCGCCCCCGGGUGGACUCGAACCACCAA-CCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUCGUACAUUCUGUUAGACUAA------- ----..((((((.......--.....(((.((((((((.......)))))..-...((((((.....)))))).((((.........))))..))).)))............))))))...------- ( -36.65, z-score = -3.17, R) >droSim1.chr2R 935253 114 + 19596830 ----GCAUCUAAAAAAGUA--AAUUCGCCCCCGGGUGGACUCGAACCACCAA-CCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUUAUGCAUUUCAUUCGACUAA------- ----((((........(..--....)(((.((((((((.......)))))..-...((((((.....)))))).((((.........))))..))).)))..))))...............------- ( -32.30, z-score = -2.22, R) >anoGam1.chr2L 44929514 93 + 48795086 -----AAUAAAAGAGUAUUU-CCUGCGCCCCCGGGUGGACUCGAACCACCAA-CCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGAAGGC---------------------------- -----.........(((...-..)))(((..(((((((.......)))))..-...((((((.....)))))).((((.........))))..))..)))---------------------------- ( -25.70, z-score = -0.77, R) >consensus ____GCGUAAAAAAAUUUA__AGCUCGCCCCCGGGUGGACUCGAACCACCAA_CCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUGAUGCAUGUUGUUGGACUAA_______ ..........................(((.((((((((.......)))))......((((((.....)))))).((((.........))))..))).)))............................ (-29.20 = -29.45 + 0.25)
| Location | 2,040,089 – 2,040,202 |
|---|---|
| Length | 113 |
| Sequences | 12 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 75.39 |
| Shannon entropy | 0.55069 |
| G+C content | 0.53433 |
| Mean single sequence MFE | -42.44 |
| Consensus MFE | -33.16 |
| Energy contribution | -33.03 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997099 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
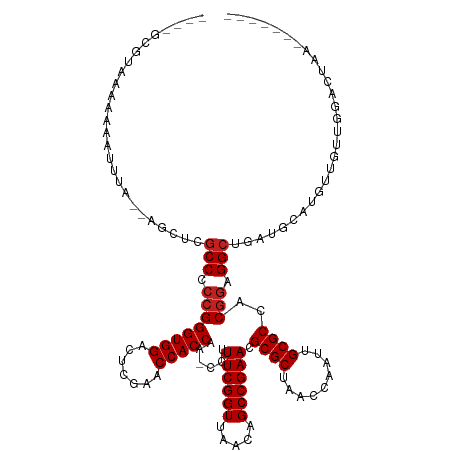
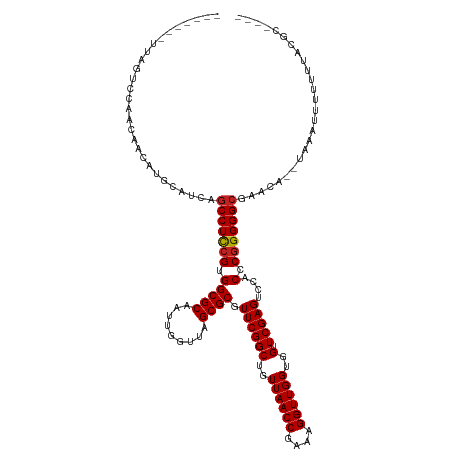
>dm3.chr2R 2040089 113 - 21146708 -------UUAGUCCAUCAAAAUGUAGAAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGG-UUGGUGGUUCGAGUCCACCCGGGGGCGACUU--UUUUAUUUU-GAUGC---- -------......((((((((((.((..(((((((.(((((.........)))).((((((..((((((....))-))))..).)))))....).)))))))..)).--...))))))-)))).---- ( -47.40, z-score = -4.32, R) >droGri2.scaffold_15245 208578 120 - 18325388 UUAGUAUUUCUUCCCAGAACUCCCAGCAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGG-UUGGUGGUUCGAGUCCACCCGGGGGCGAACU--CAACUUUUU-AUCGC---- ................((((..(((((.(((.....)))....(((((((((((.....)).)))))))))...)-))))..))))((((.(.((....)).).)))--)........-.....---- ( -40.30, z-score = -1.14, R) >droMoj3.scaffold_6496 25253680 122 - 26866924 ------UUAGAUCAUGAAAUAAACAGCAGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUUGGUGGUUCGAGUCCACCCCGGGGGGCGCGGGCAAACUUUUUUGCAAAUUC ------...................((.((((((.(((.....(((((((((((.....)).))))))))).......(((((.......))))))))))))))))..(((((....)))))...... ( -41.70, z-score = -0.84, R) >droWil1.scaffold_180700 707023 114 + 6630534 -------UUUGCCCGACCUCAGACAUGUGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGG-UUGGUGGUUCGAGUCCACCCGGGGGCGUGAG--UAAUAUUUUUUGCUU---- -------...(((((((((......((((((.....)))))).(((((((((((.....)).))))))))).)))-))(((((.......)))))...))))..(((--(((......))))))---- ( -47.90, z-score = -2.90, R) >droPer1.super_73 175288 114 + 267513 -------UAUGAAGUGAAAGCGGCAACGGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGG-UUGGUGGUUCGAGUCCACCCGGGGGCG-GCA---CAACUUUUUUACAUUU-- -------......(((((((((....))(((((((.(((((.........)))).((((((..((((((....))-))))..).)))))....).))))))).-...---.....)))))))....-- ( -44.50, z-score = -1.90, R) >dp4.chr3 17964291 114 - 19779522 -------UUUGAAGUGAAAGCGGCAACGGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGG-UUGGUGGUUCGAGUCCACCCGGGGGCG-GCA---CAAUUUUUUUACAUUU-- -------......(((((((((....))(((((((.(((((.........)))).((((((..((((((....))-))))..).)))))....).))))))).-...---.....)))))))....-- ( -44.50, z-score = -2.05, R) >droAna3.scaffold_13266 1127335 116 + 19884421 -------UUUGGCAAGCUCGAAGCAUGUGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGG-UUGGUGGUUCGAGUCCACCCGGGGGCG-GCG-CUUCUCUUUUUUGUGUGU-- -------....(((((...(((((...((((((((.(((((.........)))).((((((..((((((....))-))))..).)))))....).))))))))-..)-)))).....)))))....-- ( -45.10, z-score = -1.06, R) >droYak2.chr2L 1886453 104 + 22324452 -------UUAGACCAAGGUCGCAUGUAAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGG-UUGGUGGUUCGAGUCCACCCGGGGGCG---A--UUUAUUUU----------- -------...(((....)))........(((((((.(((((.........)))).((((((..((((((....))-))))..).)))))....).))))))).---.--........----------- ( -40.90, z-score = -2.30, R) >droEre2.scaffold_4929 6348498 114 + 26641161 -------UUAGACCAAGAACUUAUGUCAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGG-UUGGUGGUUCGAGUCCACCCGGGGGCGAAAA--ACAAUUUUUUGAUCC---- -------...((.((((((......((.(((((((.(((((.........)))).((((((..((((((....))-))))..).)))))....).)))))))))...--.....)))))).)).---- ( -40.14, z-score = -1.93, R) >droSec1.super_26 682912 114 - 826190 -------UUAGUCUAACAGAAUGUACGAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGG-UUGGUGGUUCGAGUCCACCCGGGGGCGAAUU--UGAUUUUUUAGACGC---- -------...((((((.(((((......(((((((.(((((.........)))).((((((..((((((....))-))))..).)))))....).))))))).....--..)))))))))))..---- ( -44.12, z-score = -2.83, R) >droSim1.chr2R 935253 114 - 19596830 -------UUAGUCGAAUGAAAUGCAUAAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGG-UUGGUGGUUCGAGUCCACCCGGGGGCGAAUU--UACUUUUUUAGAUGC---- -------...............((((..(((((((.(((((.........)))).((((((..((((((....))-))))..).)))))....).))))))).....--..((.....))))))---- ( -38.60, z-score = -1.53, R) >anoGam1.chr2L 44929514 93 - 48795086 ----------------------------GCCUUCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGG-UUGGUGGUUCGAGUCCACCCGGGGGCGCAGG-AAAUACUCUUUUAUU----- ----------------------------(((((((.(((((.........)))).((((((..((((((....))-))))..).)))))....).))))))).....-...............----- ( -34.10, z-score = -1.20, R) >consensus _______UUAGUCCAACAACAUGCAUCAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGG_UUGGUGGUUCGAGUCCACCCGGGGGCGAACA__UAAAUUUUUUUACGC____ ............................(((((((........(((((((((((.....)).))))))))).......(((((.......)))))))))))).......................... (-33.16 = -33.03 + -0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:04 2011