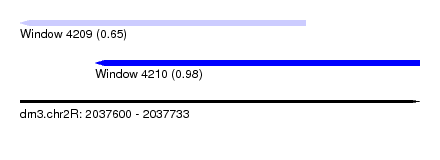
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,037,600 – 2,037,733 |
| Length | 133 |
| Max. P | 0.982555 |

| Location | 2,037,600 – 2,037,695 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.72 |
| Shannon entropy | 0.48116 |
| G+C content | 0.52651 |
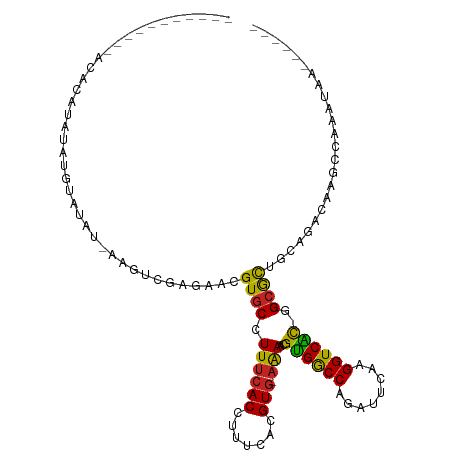
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -14.39 |
| Energy contribution | -13.99 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.649583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
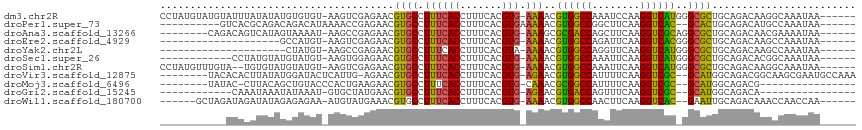
>dm3.chr2R 2037600 95 - 21146708 GUGCCUUUCACCUUUCACGUG-AAAACGUGGCCAAAUCCAAGGUCAUGGGCGCUGCAGACAAGGCAAAUAAGAGGCCAAAACCCCCAGGACACCCA--------- (((((((((((.......)))-))).(((((((........)))))))))))).........(((.........)))..........((....)).--------- ( -26.90, z-score = -0.65, R) >droPer1.super_73 173558 96 + 267513 GUGCCUUUCACCUUUCACGUGAAAAACGUGGCCGGCUUCAAGGUCACG--CACUGCAGACAUGCCAAAUAAAAGCUGCCACCACCCCCCAAACACCAA------- ((((.((((((.......))))))...((((((........)))))))--))).((((.(.............)))))....................------- ( -22.22, z-score = -1.15, R) >dp4.chr3 17962535 96 - 19779522 GUGCCUUUCACCUUUCACGUGAAAAACGUGGCCGGCUUCAAGGUCACG--CACUGCAGACAUGCCAAAUAAAAGCUGCCACCACCCCCCAAACACCAA------- ((((.((((((.......))))))...((((((........)))))))--))).((((.(.............)))))....................------- ( -22.22, z-score = -1.15, R) >droAna3.scaffold_13266 1124842 94 + 19884421 GUGCCUUUCACCUUUCACGUG-AAAGCGCGACCAGCUUCAAGGUCGCAGGCGCUGCAGACAACGAAAAUAAAAGGCCAAGA-ACCCGGGCCACCCA--------- (((((((((((.......)))-)))..((((((........)))))).)))))....................((((....-.....)))).....--------- ( -31.00, z-score = -1.99, R) >droEre2.scaffold_4929 6346163 104 + 26641161 GUGCCUUUCACCUUUCACGUG-AAAACGUGGCCAGAUUCAAGGUCACGGGCGCUGCAGACAAGCCAAAUAAAAGGCCAAAACCCCCAGGACACACAGGACACCCA (((((((((((.......)))-))).(((((((........)))))))))))).........(((........))).....((....)).......((....)). ( -29.00, z-score = -1.17, R) >droYak2.chr2L 1884245 84 + 22324452 GUGCCUUUCACCUUUCACGUA-AAAACGUGGCCAGGUUCAAGGUCAUGGGCGCUGCAGACAAGCCAAAUAAAAGGUGGUGGCCCA-------------------- (.(((..((((((((......-....(((((((........)))))))(((..((....)).))).....)))))))).))).).-------------------- ( -28.70, z-score = -1.38, R) >droSec1.super_26 680676 95 - 826190 GUGCCUUUCACCUUUCACGUG-AAAACGUGGCCAAAUUCAAGGUCAUGGGCGCUGCAGACACGGCAAAUAAAAGGCCAAAACCCCCAGGACACCCA--------- (((((((((((.......)))-))).(((((((........)))))))))))).........(((.........)))..........((....)).--------- ( -27.00, z-score = -0.90, R) >droSim1.chr2R 932975 95 - 19596830 GUGCCUUUCACCUUUCACGUG-AAAACGUGGCCAAAUUCAAGGUCAUGGGCGCUGCAGACAAGGCAAAUAAGAGGCCAAAACCCCCAGGACACCCA--------- (((((((((((.......)))-))).(((((((........)))))))))))).........(((.........)))..........((....)).--------- ( -26.90, z-score = -0.72, R) >droWil1.scaffold_180700 5926307 76 - 6630534 GUGCCUUUCACCUUUCACGUG-AAAACGUGGCCAACUUCAAGGUCACGA--AUUGCAGACAAACCAACCAAAAAGUUGG-------------------------- .(((.((((((.......)))-))).(((((((........))))))).--...)))......(((((......)))))-------------------------- ( -22.20, z-score = -2.67, R) >consensus GUGCCUUUCACCUUUCACGUG_AAAACGUGGCCAGAUUCAAGGUCACGGGCGCUGCAGACAAGCCAAAUAAAAGGCCAAAACCCCCAGGACACCCA_________ ..(((((((((.......))).....(((((((........)))))))......................))))))............................. (-14.39 = -13.99 + -0.40)
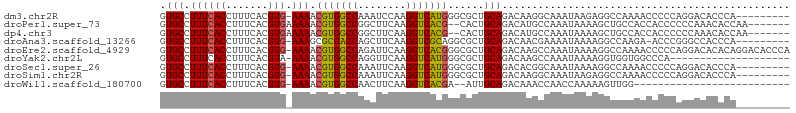
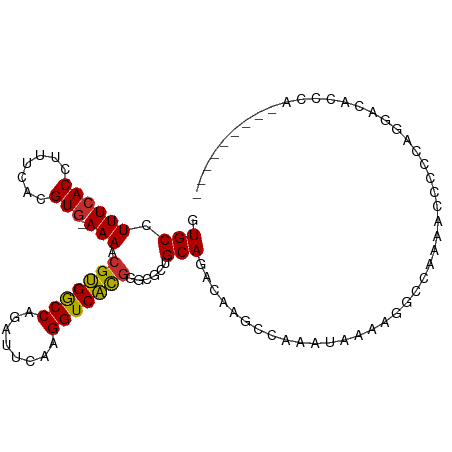
| Location | 2,037,625 – 2,037,733 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.51 |
| Shannon entropy | 0.65993 |
| G+C content | 0.47336 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -13.36 |
| Energy contribution | -12.13 |
| Covariance contribution | -1.23 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2037625 108 - 21146708 CCUAUGUAUGUAUUUAUAUAUGUGUGU-AAGUCGAGAACGUGCCUUUCACCUUUCACGUG-AAAACGUGGCCAAAUCCAAGGUCAUGGGCGCUGCAGACAAGGCAAAUAA------ ((((((((((....)))))))((.(((-(.((.....))(((((((((((.......)))-))).(((((((........)))))))))))))))).)).))).......------ ( -31.40, z-score = -1.94, R) >droPer1.super_73 173585 98 + 267513 ----------GUCACGCAGACAGACAUAAAACCGAGAACGUGCCUUUCACCUUUCACGUGAAAAACGUGGCCGGCUUCAAGGUCAC--GCACUGCAGACAUGCCAAAUAA------ ----------(((..((((.........................((((((.......))))))..(((((((........))))))--)..)))).)))...........------ ( -24.10, z-score = -0.82, R) >droAna3.scaffold_13266 1124866 100 + 19884421 --------CAGACAGUCAUAGUAAAAU-AAGCCGAGAACGUGCCUUUCACCUUUCACGUG-AAAGCGCGACCAGCUUCAAGGUCGCAGGCGCUGCAGACAACGAAAAUAA------ --------......(((.((((.....-..(((........((.((((((.......)))-)))))((((((........)))))).)))))))..)))...........------ ( -26.61, z-score = -1.39, R) >droEre2.scaffold_4929 6346197 88 + 26641161 --------------------GCCAUGU-AAGUCGAGAACGUGCCUUUCACCUUUCACGUG-AAAACGUGGCCAGAUUCAAGGUCACGGGCGCUGCAGACAAGCCAAAUAA------ --------------------.......-..(((..(...(((((((((((.......)))-))).(((((((........))))))))))))..).)))...........------ ( -27.10, z-score = -1.71, R) >droYak2.chr2L 1884259 87 + 22324452 ---------------------CUAUGU-AAGCCGAGAACGUGCCUUUCACCUUUCACGUA-AAAACGUGGCCAGGUUCAAGGUCAUGGGCGCUGCAGACAAGCCAAAUAA------ ---------------------...(((-..((.(.(..((((((((..((((.((((((.-...))))))..))))..)))).))))..).).))..)))..........------ ( -26.10, z-score = -1.59, R) >droSec1.super_26 680701 96 - 826190 ------------CCUAUGUAUGUAUGU-AAGUGGAGAACGUGCCUUUCACCUUUCACGUG-AAAACGUGGCCAAAUUCAAGGUCAUGGGCGCUGCAGACACGGCAAAUAA------ ------------....(((.(((.(((-(.((.....))(((((((((((.......)))-))).(((((((........)))))))))))))))).)))..))).....------ ( -28.50, z-score = -1.40, R) >droSim1.chr2R 933000 106 - 19596830 CCUAUGUUUGUA--UGUGUAUGUAUGU-AAGUCGAGAACGUGCCUUUCACCUUUCACGUG-AAAACGUGGCCAAAUUCAAGGUCAUGGGCGCUGCAGACAAGGCAAAUAA------ ....(((((((.--..(((.((((...-..((.....))(((((((((((.......)))-))).(((((((........)))))))))))))))).)))..))))))).------ ( -32.90, z-score = -2.23, R) >droVir3.scaffold_12875 4763625 104 - 20611582 --------UACACACUUAUAUGGAUACUCAUUG-AGAACGUGCCUUUCACCUUUCACGUG-AGAACGUGGCCAUUUUCAAGGUCGC--GCAUGGCAGACGGCAAGCGAAUGCCAAA --------......((((.((((....))))))-))....((((((((((.......)))-))).(((((((........))))))--)...))))...((((......))))... ( -28.70, z-score = -1.23, R) >droMoj3.scaffold_6496 1562486 88 + 26866924 --------UAUAC-CUUACAGCUGUACCCACUGAAGAACGUGCCUUUCACCUUUCACGUG-CAAACGCGGCCAUUUUCAAGGUCGC--GCAUGGCAGACG---------------- --------.....-......(((((...((((((((...(((.....))).))))).)))-((..(((((((........))))))--)..)))))).).---------------- ( -22.40, z-score = -1.08, R) >droGri2.scaffold_15245 18104193 84 + 18325388 ------------CAAAUAAAUAUAAAU-GUGCUAUGAACGUGCCUUUCACCUUUCACGUG-AGAACGUGACCAGUUUCAAGGUCGC--GCAUGGCAGACA---------------- ------------..............(-.(((((((..((((((((..((...((((((.-...))))))...))...)))).)))--)))))))).)..---------------- ( -24.80, z-score = -2.49, R) >droWil1.scaffold_180700 5926315 100 - 6630534 ------GCUAGAUAGAUAUAGAGAGAA-AUGUAUGAAACGUGCCUUUCACCUUUCACGUG-AAAACGUGGCCAACUUCAAGGUCAC--GAAUUGCAGACAAACCAACCAA------ ------((............(((((..-((((.....))))..)))))....(((..((.-...))((((((........))))))--)))..))...............------ ( -21.10, z-score = -1.38, R) >consensus ___________ACACAUAUAUGUAUAU_AAGUCGAGAACGUGCCUUUCACCUUUCACGUG_AAAACGUGGCCAGAUUCAAGGUCAC_GGCGCUGCAGACAAGCCAAAUAA______ .......................................((((..............((.....))((((((........))))))..))))........................ (-13.36 = -12.13 + -1.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:03 2011