| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,023,361 – 2,023,431 |
| Length | 70 |
| Max. P | 0.994477 |

| Location | 2,023,361 – 2,023,431 |
|---|---|
| Length | 70 |
| Sequences | 4 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 61.67 |
| Shannon entropy | 0.61628 |
| G+C content | 0.39126 |
| Mean single sequence MFE | -13.88 |
| Consensus MFE | -8.90 |
| Energy contribution | -9.03 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.994477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2023361 70 - 21146708 CAUAGUGUACAAAAGCAUUAGGAGCUCGGUUGGUAUUACUCCAAUACCAACAAGGCUUACAAUUGUCACU ...((((.((((.........(((((..(((((((((.....)))))))))..)))))....)))))))) ( -21.32, z-score = -3.99, R) >droEre2.scaffold_4929 6331095 59 + 26641161 -----------AAAAUAUUAGGAGCUCGUUUGGUAUUACUGCAAUGCCAACAAGGCUUAAAAUUGUCACU -----------..........(((((...((((((((.....))))))))...)))))............ ( -12.60, z-score = -1.44, R) >droSec1.super_26 666404 66 - 826190 ----CUGUACAUAAACGUUAGAAGCUCGUUUGGUAUUACUCCAAUGCCAACAAGGCUUACAAUUGUCGCU ----..........(((....(((((...((((((((.....))))))))...))))).....))).... ( -12.60, z-score = -0.73, R) >droWil1.scaffold_180700 678981 60 + 6630534 -----UAUAUUGAGACAUUGCGGG-----AAGCAAAAAUUCAAAUGCCUCAACACCUCGAUAUUGUCCCA -----.((((((((...((((...-----..)))).........((......)).))))))))....... ( -9.00, z-score = -0.36, R) >consensus _____UGUACAAAAACAUUAGGAGCUCGUUUGGUAUUACUCCAAUGCCAACAAGGCUUAAAAUUGUCACU ..............(((....(((((...((((((((.....))))))))...))))).....))).... ( -8.90 = -9.03 + 0.13)


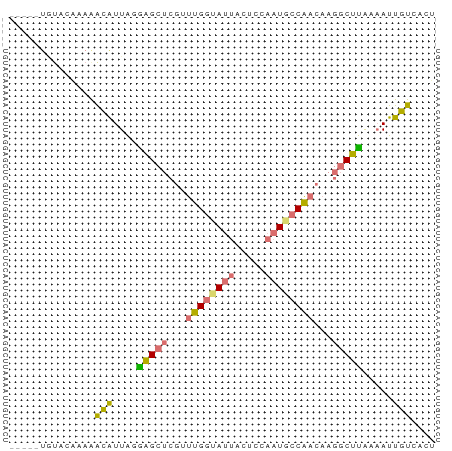
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:00 2011