| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,176,626 – 2,176,733 |
| Length | 107 |
| Max. P | 0.938912 |

| Location | 2,176,626 – 2,176,733 |
|---|---|
| Length | 107 |
| Sequences | 10 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 76.96 |
| Shannon entropy | 0.45850 |
| G+C content | 0.44562 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -13.63 |
| Energy contribution | -13.63 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.938912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
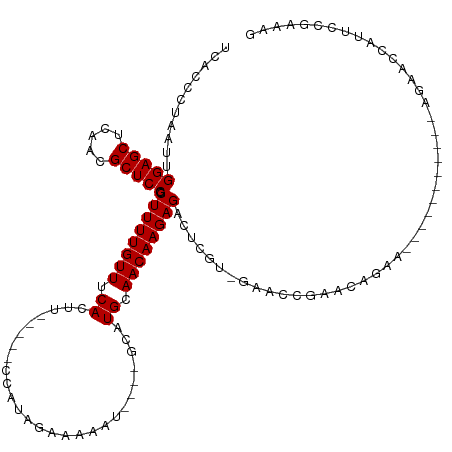
>dm3.chr2L 2176626 107 + 23011544 UCACCCUAAUUGGAGCUCAACGCUCCGCUUUUGUUUCACUUCACUUCCAUAGAAAAGU----GCAUGCAACAAGAGACUCGU-GAACCGAACAGAA-----------AGAACGAUUCCGAAAG ...........(((((.....))))).((((((((.((...(((((........))))----)..)).))))))))..(((.-(((.((.......-----------....)).))))))... ( -23.80, z-score = -1.65, R) >droSim1.chr2L 2128950 107 + 22036055 UCACCCUAAUUGGAGCUCAACGCUCCGCUUUUGUUUCACUUCACUUCCAUAGAAAAGU----GCAUGCAACAAGAGACUCGU-GAACAGAACCGAA-----------AGAACCAUUCCGAAAG ((((.......(((((.....))))).((((((((.((...(((((........))))----)..)).))))))))....))-)).......(...-----------.).............. ( -24.50, z-score = -2.29, R) >droSec1.super_121 49114 102 + 59987 UCACCCUAAUUGGAGCUCAACGCUCCGCUUUUGUUUCACUU-----CCAUAGAAAAGU----GCAUGCAACAAGAGACUCGU-GAACCGAACCGAA-----------AGAACCAUUCCGAAAG ((((.......(((((.....))))).((((((((.(((((-----........))))----).....))))))))....))-)).......(...-----------.).............. ( -21.80, z-score = -1.76, R) >droYak2.chr2L 2151106 107 + 22324452 UCACCCUAAUUGGAGCUCAACGCUCCGCUUUUGUUUCACUUCACUUCCAUAGAAAAGU----GCAUGCAACAAGAGACUCGU-GAACCGAACAGAA-----------AGAACCAUUCCGAAAG ((((.......(((((.....))))).((((((((.((...(((((........))))----)..)).))))))))....))-))...........-----------................ ( -23.50, z-score = -1.95, R) >droEre2.scaffold_4929 2213246 107 + 26641161 UCACCCUAAUUGGAGCUCAACGCUCCGCUUUUGUUUCACUUCACUUCCAUAGAAAAGU----GCAUGCAACAAGAGACUCGU-GAACCGAACAGAA-----------GGAACCAUUCCGAAAG ((((.......(((((.....))))).((((((((.((...(((((........))))----)..)).))))))))....))-))...........-----------(((.....)))..... ( -25.80, z-score = -2.13, R) >droAna3.scaffold_12916 9885626 119 - 16180835 UCACCCUAAUUGGAGCUCAACGCUCCGCUUUUGUUUCACUUCGAUUCCAUAGAAAAAU---UGCAUGCAACAAGAGACUCGU-GAACCGAACAGAACGAAGGAGAACAGAACCAUUCCAAAUG ...........(((((.....)))))(.((((((((..(((((.(((..........(---((....)))........(((.-....)))...))))))))..)))))))).).......... ( -25.80, z-score = -1.69, R) >droWil1.scaffold_180708 10449192 91 - 12563649 GCACCCUAAUUGGAGCUUAACGCUCCGCUUUUGUUUCACUU-----CCAUAGAAAAA--AGUGCAUGCAACAAGAGACUCGUCGAACCCAA-----------CCCAAAU-------------- ...........(((((.....))))).((((((((.(((((-----..........)--)))).....))))))))...............-----------.......-------------- ( -16.40, z-score = -1.30, R) >droVir3.scaffold_12963 11731995 89 - 20206255 UCAACCCAAUUGGAGCUUAACGCUCCGCUUUUGUUUCACUU-----CCAUAGAAAAA--AGUGCAUGCAACAAGAGACCCGUCGAACCCAA---AG------CAA------------------ ...........(((((.....)))))((((((((..(((((-----..........)--))))...)))......((....))......))---))------)..------------------ ( -17.30, z-score = -1.49, R) >droMoj3.scaffold_6500 31269462 109 - 32352404 UCAAUCCAAUUGGAGCUUAACGCUCCGCUUUUGUUUCACUU-----CCAUAGAAAAAUUGCUGCAUGCAACAAGAGACCCGUCGAACCCAG---AG------CCCAAAAACCCAACGCCCAAA ...........(((((.....)))))(((((.(.(((....-----...........((((.....)))).....((....))))).).))---))------).................... ( -19.00, z-score = -1.35, R) >droGri2.scaffold_15252 5534261 106 - 17193109 ----UCCAAUUGGAGCUUAACGCUCCGCUUUUGUUACACUU-----CCAUAGAAAAA--AGUGCAUGCAACAAGAGAAGCGUCGAACCAAACUAAA------UAAAAAGAACAAAUGCAACAG ----.......(((((.....)))))((.((((((.(((((-----..........)--)))).((((..(....)..))))..............------.......)))))).))..... ( -19.40, z-score = -1.43, R) >consensus UCACCCUAAUUGGAGCUCAACGCUCCGCUUUUGUUUCACUU_____CCAUAGAAAAAU____GCAUGCAACAAGAGACUCGU_GAACCGAACAGAA___________AGAACCAUUCCGAAAG ...........(((((.....))))).((((((((.((...........................)).))))))))............................................... (-13.63 = -13.63 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:27 2011