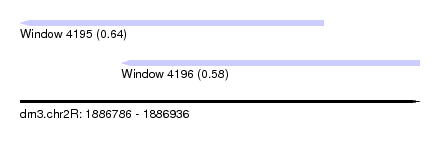
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,886,786 – 1,886,936 |
| Length | 150 |
| Max. P | 0.642764 |

| Location | 1,886,786 – 1,886,900 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.11 |
| Shannon entropy | 0.50081 |
| G+C content | 0.53289 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -20.53 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
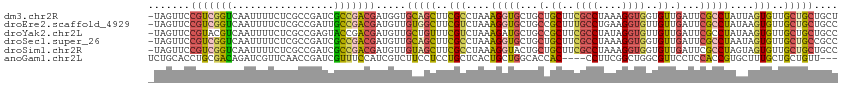
>dm3.chr2R 1886786 114 - 21146708 -UAGUUCCGUCGGUCAAUUUUCUCGCCGAUCGCCGACGAUGGUGCAGCUUCGCCUAAAGGUGCUGCUGCUUCGCCUAAAGGUGGUGUUGAUUCGCCUAUUAGUGUUGCUGCUGCU -......(((((((..(((........))).)))))))..((((((((..(((.((((((((...(.((.(((((....))))).)).)...))))).))))))..))))).))) ( -37.10, z-score = -0.91, R) >droEre2.scaffold_4929 6193566 114 + 26641161 -UAGUUCCGUCGGUCAAUUUUCUCGCCGAUUGCCGACGAUGUUGUGGCUUCGUCUAAAGGUGCUGCCGCUUUGCCUGAAGGUGUUGUUGAUUCGCCUAUAAGUGUUGCUGCUGCC -......((((((.(((((........)))))))))))..((.(..((..((..((.(((((...(.((..((((....))))..)).)...))))).))..))..))..).)). ( -35.10, z-score = -1.86, R) >droYak2.chr2L 1707829 114 + 22324452 -UAGUUCCGUACGUCAAUUUUCUCGCCGAGUACCGACGAUGUUGCUGUUUCGUCUAAAGAUGCUGCCGCUUCGCCUAUAGGUGUUGUUGAUUCGCCUAUAAGUGUUGCUGCUGCC -((((..(((.((((....(((.....)))....)))))))..))))....(((....)))((.((.((..(((.((((((((.........)))))))).)))..)).)).)). ( -31.50, z-score = -1.78, R) >droSec1.super_26 532013 114 - 826190 -UAGUUCCGUCGGUCAAUUUUCUCGCCGAUCGCCGACGAUGUUGCAGCUUCGCCUAAAGGUGCUGCUGCUUCGCCUAAAGGUGGUGUUGAUUCGCCUAAUAGUGUUGCUGCCGCC -......(((((((..(((........))).)))))))..((.(((((..(((....(((((...(.((.(((((....))))).)).)...)))))....)))..))))).)). ( -36.80, z-score = -1.33, R) >droSim1.chr2R 787924 114 - 19596830 -UAGUUCCGUCGGUCAAUUUUCUCGCCGAUCGCCGACGAUGUUGUAGCUUCGCCUAAAGGUACUGCUGCUUCGCCUAAAGGUGGUGUUGAUUCGCCUAGUAGUGUUGCUGCUGCC -......(((((((..(((........))).)))))))..((.(((((...(((....)))(((((((....(((....)))((((......)))))))))))...))))).)). ( -33.70, z-score = -0.83, R) >anoGam1.chr2L 20397819 108 + 48795086 UCUGCACCUGCGACAGAUCGUUCAACCGAUCGUUUCCAUCGUCUUCCUCCUGCUCACUGCUGGCACCAC----CCUUCGGCUGGCGUUCCUCCACCGUGCUUUGCUGCUGUU--- ...((((....(((.(((((......)))))(.......))))...............((..((.....----......))..))...........))))............--- ( -19.80, z-score = 1.23, R) >consensus _UAGUUCCGUCGGUCAAUUUUCUCGCCGAUCGCCGACGAUGUUGCAGCUUCGCCUAAAGGUGCUGCCGCUUCGCCUAAAGGUGGUGUUGAUUCGCCUAUUAGUGUUGCUGCUGCC .......(((((((.................))))))).....(((((..(((....(((((...(.((..((((....))))..)).)...)))))....)))..))))).... (-20.53 = -20.90 + 0.37)

| Location | 1,886,824 – 1,886,936 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.04 |
| Shannon entropy | 0.56083 |
| G+C content | 0.53159 |
| Mean single sequence MFE | -33.91 |
| Consensus MFE | -13.42 |
| Energy contribution | -14.91 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.583888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
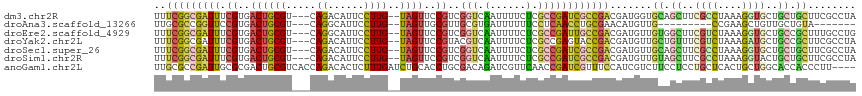
>dm3.chr2R 1886824 112 - 21146708 UUUCGGCGAUUUCGUGACUGCGU---CAGACAUUCCUUG--UAGUUCCGUCGGUCAAUUUUCUCGCCGAUCGCCGACGAUGGUGCAGCUUCGCCUAAAGGUGCUGCUGCUUCGCCUA ..(((((((...((..((((((.---...........))--))))..))(((((..........))))))))))))....((((((((..((((....))))..)))))...))).. ( -41.70, z-score = -2.66, R) >droAna3.scaffold_13266 979254 96 + 19884421 UUGCGCCGGUUCCGUGACUGCGU---CAGGCAUUCCUUG--UAGUUGCGUUGCGUGAUUUUUUCCUCAACCUGCGAACAUGUUG---------CCGAAGCUGUUGCUGUA------- .(((....((..((..((((((.---.(((....)))))--))))..))..))(..((..((((..((((.((....)).))))---------..))))..))..).)))------- ( -25.90, z-score = -0.04, R) >droEre2.scaffold_4929 6193604 112 + 26641161 UUUCGGCGAUUUCGUGACUGCGU---CAGGCAUUCCUUG--UAGUUCCGUCGGUCAAUUUUCUCGCCGAUUGCCGACGAUGUUGUGGCUUCGUCUAAAGGUGCUGCCGCUUUGCCUG ....(((((..(((..((((((.---.(((....)))))--))))..)(((((.(((((........))))))))))))....(((((..(..(....)..)..))))).))))).. ( -37.10, z-score = -1.72, R) >droYak2.chr2L 1707867 112 + 22324452 UUUCGGCGAUUUCGUGACUGCGU---CAGACAUUCCUUG--UAGUUCCGUACGUCAAUUUUCUCGCCGAGUACCGACGAUGUUGCUGUUUCGUCUAAAGAUGCUGCCGCUUCGCCUA .((((((((.....((((((((.---...(((.....))--).....)))).))))......))))))))....(.(((....((.((..((((....))))..)).)).))).).. ( -31.00, z-score = -1.12, R) >droSec1.super_26 532051 112 - 826190 UUUCGGCGAUUUCGUGACUGCGU---CAGACAUUCCUUG--UAGUUCCGUCGGUCAAUUUUCUCGCCGAUCGCCGACGAUGUUGCAGCUUCGCCUAAAGGUGCUGCUGCUUCGCCUA ..(((((((...((..((((((.---...........))--))))..))(((((..........))))))))))))(((....(((((..((((....))))..))))).))).... ( -41.50, z-score = -3.08, R) >droSim1.chr2R 787962 112 - 19596830 UUUCGGCGAUUUCGUGACUGCGU---CAGACAUUCCUUG--UAGUUCCGUCGGUCAAUUUUCUCGCCGAUCGCCGACGAUGUUGUAGCUUCGCCUAAAGGUACUGCUGCUUCGCCUA ..(((((((...((..((((((.---...........))--))))..))(((((..........))))))))))))(((....(((((...(((....)))...))))).))).... ( -36.40, z-score = -2.15, R) >anoGam1.chr2L 20397854 113 + 48795086 UUGCGCCGAUUGCGCGACUGCGUCACCAGACACUCUUUGAUCUGCACCUGCGACAGAUCGUUCAACCGAUCGUUUCCAUCGUCUUCCUCCUGCUCACUGCUGGCACCACCCUU---- ((((((.....))))))....((((.(((.........(((((((......).)))))).......((((.......))))...............))).)))).........---- ( -23.80, z-score = -0.00, R) >consensus UUUCGGCGAUUUCGUGACUGCGU___CAGACAUUCCUUG__UAGUUCCGUCGGUCAAUUUUCUCGCCGAUCGCCGACGAUGUUGCAGCUUCGCCUAAAGGUGCUGCUGCUUCGCCUA ....(((((....(((.(((......))).)))..............(((((((.((((........))))))))))).....((.((..((((....))))..)).)).))))).. (-13.42 = -14.91 + 1.50)
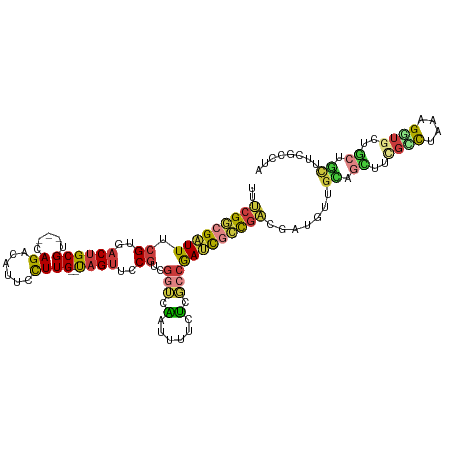
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:51 2011