
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,885,281 – 1,885,399 |
| Length | 118 |
| Max. P | 0.801813 |

| Location | 1,885,281 – 1,885,399 |
|---|---|
| Length | 118 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.65 |
| Shannon entropy | 0.55310 |
| G+C content | 0.57117 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -22.43 |
| Energy contribution | -22.35 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.32 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1885281 118 + 21146708 --UAAUCUGCACAAGGAACCAUCAUGACGGCCAUGCCCGUGGUGUGGUGCACCGAUCCCAACUCGGUGAACAAUAAGCCCACAGCCUUCGCCUUGCACGGAAUUCAGCUGCAAGGAAAUG --..........((((.((((.(((.((((......)))).))))))).((((((.......))))))................))))..((((((((........).)))))))..... ( -34.80, z-score = -0.48, R) >droGri2.scaffold_15245 8916904 110 + 18325388 ----------CAAUCCAAACAAGAUGACCACCAUGCCAGUGGUUUGGUGCACCGAUCCCAACUCGGUGAACAACAAACCCACAGCCUUUGCCCUGCACGGCAUACAGCUGCAGGGUAAUG ----------........................((..((((((((...((((((.......)))))).....)))).)))).))..((((((((((..((.....)))))))))))).. ( -34.70, z-score = -1.84, R) >droMoj3.scaffold_6496 3231083 110 + 26866924 ----------UAAUCCGAGCAAGAUGACCACCAUGCCCGUGGUCUGGUGCACCGAUCCCAAUUCGGUGAACAACAAGCCAACGGCCUUUGCUCUGCAUGGCAUACAGCUGCAGGGCAAUG ----------....(((........((((((.......))))))((((.((((((.......))))))........)))).)))...((((((((((..((.....)))))))))))).. ( -39.60, z-score = -1.92, R) >droVir3.scaffold_12823 1719877 110 - 2474545 ----------CAUUCCCAUCAGGAUGACCACCAUGCCGGUGGUGUGGUGCACCGAUCCAAAUUCGGUGAACAACAAACCAACUGCGUUUGCCCUGCACGGCAUACAGCUGCAGGGCAAUG ----------(((((......)))))((((((.....)))))).((((.((((((.......))))))........)))).......((((((((((..((.....)))))))))))).. ( -43.10, z-score = -2.41, R) >dp4.chr3 8093136 120 - 19779522 GCAUCCGCCCAGAGCCUGCCAGCAUGACUGCCAUGCCCGUGGUGUGGUGCACCGAUCCGAAUUCGGUGAACAACAAGCCCACGGCCUUCGCCCUGCACGGCAUCCAGCUGCAGGGCAACG (((...((.....)).)))..(((((.....)))))(((((((((.((.((((((.......)))))).)).)))...)))))).....((((((((..((.....)))))))))).... ( -47.20, z-score = -1.15, R) >droAna3.scaffold_13266 977722 118 - 19884421 -CAGCCACCGA-AGCGAUCCAUCAUGACAGCCAUGCCCGUCGUGUGGUGCACCGACCCCAACUCGGUGAACAACAAGCCCACAGCCUUCGCCCUUCACGGCAUCCAGUUGCAGGGCAAUG -..(((((...-.(((((....((((.....))))...)))))))))).((((((.......))))))........((((.((((....(((......))).....))))..)))).... ( -35.30, z-score = -0.79, R) >droEre2.scaffold_4929 6192064 118 - 26641161 --UAAUCUGCACAAGGAACCAUCAUGACGGCCAUGCCCGUGGUUUGGUGCACCGAUCCCAACUCGGUGAACAAUAAGCCCACGGCCUUCGCCCUGCACGGAAUCCAGCUGCAAGGGAAUG --..........((((......((((.....)))).((((((((((...((((((.......)))))).....)))).))))))))))..((((((((........).))).)))).... ( -33.60, z-score = 0.51, R) >droYak2.chr2L 1706330 118 - 22324452 --UAAUUUGCUCAAGGAACCAUCAUGACGGCCAUGCCCGUGGUGUGGUGCACUGAUCCCAACUCGGUAAACAAUAAGCCCACGGCCUUCGCCUUGCACGGAAUCCAGCUGCAAGGGAACG --......(((...((.((((.(((.((((......)))).)))))))..(((((.......)))))..........))...))).(((.((((((((........).)))))))))).. ( -32.00, z-score = 0.80, R) >droSec1.super_26 530515 118 + 826190 --UAAUCUGUACAAGGAGCCAUCAUGACGGCCAUGCCCGUGGUGUGGUGCACCGAUCCCAACUCGGUGAACAAUAAGCCCACCCGCUUCGCCUUGCACGGAAUUCAGCUGCAAGGAAAUG --............(((((.......((((......))))((((.(((.((((((.......))))))........))))))).))))).((((((((........).)))))))..... ( -37.30, z-score = -1.01, R) >droSim1.chr2R 786419 118 + 19596830 --UAAUCUGUACAAGGAGCCAUCAUGACGGCCAUGCCCGUGGUGUGGUGCACCGAUCCCAACUCGGUGAACAAUAAGCCCACAGCCUUCGCCUUGCACGGAAUUCAGCUGCAAGGAAAUG --....((((....((.((((.(((.((((......)))).))))))).((((((.......))))))..........))))))......((((((((........).)))))))..... ( -35.10, z-score = -0.37, R) >anoGam1.chr2L 20396242 101 - 48795086 ----------------AGCCGCCAUGUCCGCGGUACCGGUCGUCUGGUGCUCGGACCCGAACUCGUCGGAAAAGAAGCA---ACCGUUCGCGCUGCAGGGCAUCCAGCUGCAGGGCAACA ----------------....((...(((((.((((((((....)))))))))))))((((.....)))).......)).---...(((.((.((((((.........)))))).))))). ( -40.60, z-score = -0.57, R) >droWil1.scaffold_180700 4684064 97 - 6630534 -----------------------AUGACUGCCAUACCCGUGGUAUGGUGUACCGAUCCCAAUUCGGUUAAUAAUAAGCCCACAGCAUUUGCUCUCCAUGGCAUUCAGUUGCAGGGGAAUG -----------------------......((((((((...))))))))..(((((.......)))))..........(((.((((...((((......))))....))))...))).... ( -26.80, z-score = -0.34, R) >droPer1.super_4 73713 102 - 7162766 ------------------CCAGCAUGACUGCCAUGCCCGUGGUGUGGUGCACCGAUCCCAAUUCGGUGAACAACAAGCCCACGGCCUUCGCCCUGCACGGCAUCCAGCUGCAGGGCAACG ------------------...(((((.....)))))(((((((((.((.((((((.......)))))).)).)))...)))))).....((((((((..((.....)))))))))).... ( -45.50, z-score = -2.91, R) >consensus __________ACAACGAACCAUCAUGACGGCCAUGCCCGUGGUGUGGUGCACCGAUCCCAACUCGGUGAACAACAAGCCCACAGCCUUCGCCCUGCACGGCAUCCAGCUGCAGGGCAAUG .............................((((..(.....)..)))).((((((.......))))))......................((((((((........).)))))))..... (-22.43 = -22.35 + -0.08)
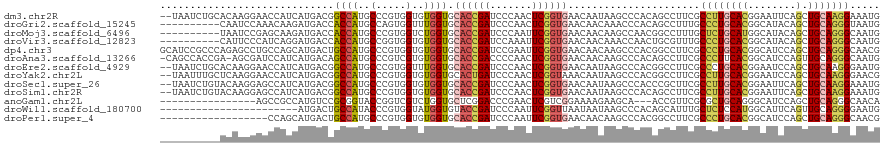

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:49 2011