| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,884,862 – 1,884,975 |
| Length | 113 |
| Max. P | 0.783512 |

| Location | 1,884,862 – 1,884,975 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.00 |
| Shannon entropy | 0.57647 |
| G+C content | 0.33576 |
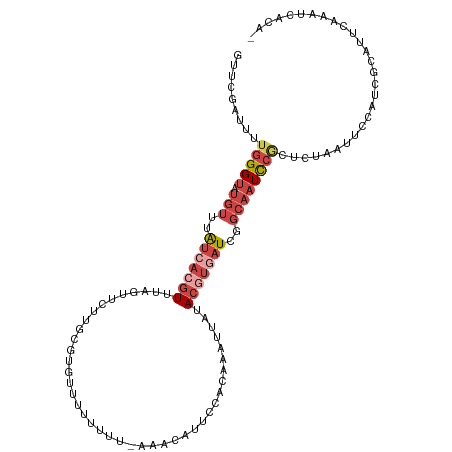
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -5.39 |
| Energy contribution | -7.11 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.20 |
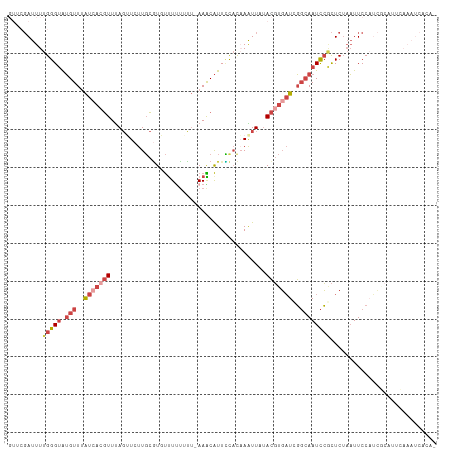
| Mean z-score | -2.54 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
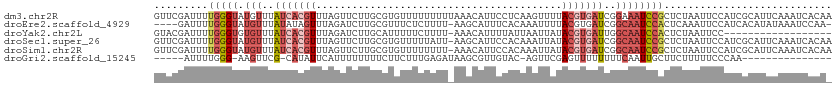
>dm3.chr2R 1884862 113 + 21146708 GUUCGAUUUUGGGUAUGUUUAUCACGUUUAGUUCUUGCGUGUUUUUUUUUAAACAUUCCUCAAGUUUUACGUGAUCGGAAAUCCGCUCUAAUUCCAUCGCAUUCAAAUCACAA ((..(((((.(((.(((((((.(((((.........)))))........))))))).))).)))))..))(((((.((((...........)))))))))............. ( -24.20, z-score = -2.90, R) >droEre2.scaffold_4929 6191626 107 - 26641161 ----GAUUUUGGGUAUGUUUAUAUAGUUUAGAUCUUGCGUUUCUCUUUU-AAGCAUUUCACAAAUUUUACGUGAUCGGCAAUCCACUCAAAUUCCAUCACAUAUAAAUCCAA- ----....((((((....((((((.(((((((..............)))-))))................(((((.((.(((........))))))))))))))))))))))- ( -15.84, z-score = -1.12, R) >droYak2.chr2L 1705958 94 - 22324452 GUACGAUUUUGGGUGUGUUUAUCACGUUUAGAUCUUGCAUUUUUCUUUU-AAACAUUUUAUUAAUUAUACGUGAUUGGCAAUCCACUCUAAUUCC------------------ ....((.((.((((((((..((((((((((((..............)))-))))....(((.....))).)))))..)))...))))).)).)).------------------ ( -16.44, z-score = -1.69, R) >droSec1.super_26 530075 112 + 826190 GUUCGAUUUUGGGUAUGUUUAUCACGUUUAGUUCUUGCGUGUUUUUAUU-AAGCAUUCCACAAAUUAUACGUGAUCGGCAAUCCGCUCUAAUUCCAUCGCAUUCAAAUCACAA ....(((((.((((.(((..(((((((.(((((..((.((((((.....-))))))..))..))))).)))))))..)))))))((............))....))))).... ( -26.80, z-score = -3.71, R) >droSim1.chr2R 785977 112 + 19596830 GUUCGAUUUUGGGUAUGUUUAUCACGUUUAGUUCUUGCGUGUUUUUUUU-AAACAUUCCACAAAUUAUACGUGAUCGGCAAUCCGCUCUAAUUCCAUCGCAUUCAAAUCACAA ....(((((.((((.(((..(((((((.(((((..((.((((((.....-))))))..))..))))).)))))))..)))))))((............))....))))).... ( -26.80, z-score = -4.05, R) >droGri2.scaffold_15245 8916452 90 + 18325388 -----AUUUUGGG-AAGUUCG-CAUAUUCAUUUUUUUUCUUCUUUGAGAUAAGCGUUGUAC-AGUUCGAGUUUUUUUUCAAUUGCUUCUUUUUCCCAA--------------- -----...(((((-(((..((-(.((((((..............))).))).))).....(-((((.(((......))))))))......))))))))--------------- ( -15.44, z-score = -1.76, R) >consensus GUUCGAUUUUGGGUAUGUUUAUCACGUUUAGUUCUUGCGUGUUUUUUUU_AAACAUUCCACAAAUUAUACGUGAUCGGCAAUCCGCUCUAAUUCCAUCGCAUUCAAAUCACA_ .........(((((.(((..(((((((.........................................)))))))..))))))))............................ ( -5.39 = -7.11 + 1.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:48 2011