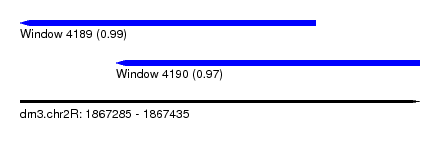
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,867,285 – 1,867,435 |
| Length | 150 |
| Max. P | 0.990529 |

| Location | 1,867,285 – 1,867,396 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.59 |
| Shannon entropy | 0.58169 |
| G+C content | 0.53056 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -20.59 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
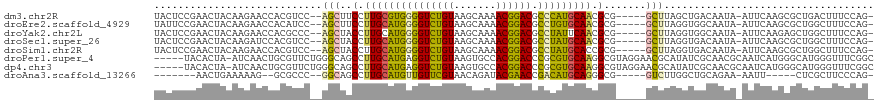
>dm3.chr2R 1867285 111 - 21146708 UACUCCGAACUACAAGAACCACGUCC--AGCUUCCUUGCGUGGGGUCUGUAAGCAAAACGGACGCCCAUGCAACGCG-----GCUUAGCUGACAAUA-AUUCAAGCGCUGACUUUCCAG- ......................(((.--((((.(.(((((((((((((((.......)))))).))))))))).).)-----)))..(((((.....-..)).)))...))).......- ( -32.30, z-score = -1.67, R) >droEre2.scaffold_4929 6174354 111 + 26641161 UAUUCCGAACUACAAGAACCACAUCC--AGCUUCCUUGCAUGGGGUCUGUAAGCAAAACGGACGCCUGUGCAACGCG-----GCUUAGGUGGCAAUA-AUUCAAGCGCUGGCUUUCCAG- ..................((((....--((((.(.(((((((((((((((.......)))))).))))))))).).)-----)))...)))).....-.........((((....))))- ( -33.60, z-score = -1.05, R) >droYak2.chr2L 1681802 111 + 22324452 UACUCCGAACUACAAGAACCACGCCC--AGCUACCUUGCAUGGGGUCUGUAAGCAAAACGGACGCCUAUUCAACGCG-----GCUUAGGUGGCAAUA-AUUCAAGAGCUGGCUUUCCAG- ......................(((.--((((...(((.(((((((((((.......)))))).))))).))).((.-----((....)).))....-.......))))))).......- ( -29.10, z-score = -0.21, R) >droSec1.super_26 512112 111 - 826190 UACUCCGAACUACAAGAUCCACGUCC--AGCUACCUUGCAUGGGGUCUGUAAGCAAAACGGACGCCUAUGCAACGCG-----GCUUAGGUGACAAUA-AUUCAAGCGCUGGCUUUCCAG- ......................(.((--(((....(((((((((((((((.......)))))).)))))))))....-----((((..((.......-))..)))))))))).......- ( -34.40, z-score = -1.93, R) >droSim1.chr2R 765963 111 - 19596830 UACUCCGAACUACAAGAACCACGUCC--AGCUACCUUGCAUGGGGUCUGUAAGCAAAACGGACGCCUAUGCACCGCG-----GCUUAGGUGACAAUA-AUUCAAGCGCUGGCUUUCCAG- ......................(.((--(((.....((((((((((((((.......)))))).)))))))).....-----((((..((.......-))..)))))))))).......- ( -33.60, z-score = -1.55, R) >droPer1.super_4 53561 114 + 7162766 -----UACACUA-AUCAACUGCGUUCUGGGCAGCCUUGCAUGAGGUCUGUAAGUGCCACGGACCCGCGUGCAAGGCGUAGGAACGCAUAUCGCAACGCAAUCAUGGGCAUGGGUUUCGGC -----.......-......((((((((..((.((((((((((.(((((((.......)))))))..)))))))))))).))))))))..(((.(((.((..........)).))).))). ( -45.20, z-score = -2.23, R) >dp4.chr3 8072713 114 + 19779522 -----UACACUA-AUCAACUGCGUUCUGGGCAGCCUUGCAUGAGGUCUGUAAGUGCCACGGACCCGCGUGCAAGGCGUAGGAACGCAUAUCGCAACGCAAUCAUGGGCAUGGGUUUCGGC -----.......-......((((((((..((.((((((((((.(((((((.......)))))))..)))))))))))).))))))))..(((.(((.((..........)).))).))). ( -45.20, z-score = -2.23, R) >droAna3.scaffold_13266 962864 97 + 19884421 -------AACUGAAAAAG--GCGCCC--GGCAGCCUUGCAUGUUGUUCGUAACAGAUACGAACCGACAUGCAGGGCG-----GUCUUGGCUGCAGAA-AAUU-----CUCGCUUCCCAG- -------..(((....((--(((((.--(((.(((((((((((((((((((.....)))))).))))))))))))).-----)))..))).(.(((.-...)-----)))))))..)))- ( -41.00, z-score = -4.09, R) >consensus UACUCCGAACUACAAGAACCACGUCC__AGCUACCUUGCAUGGGGUCUGUAAGCAAAACGGACGCCCAUGCAACGCG_____GCUUAGGUGGCAAUA_AUUCAAGCGCUGGCUUUCCAG_ .............................((....(((((((((((((((.......)))))).))))))))).))............................................ (-20.59 = -19.70 + -0.89)
| Location | 1,867,321 – 1,867,435 |
|---|---|
| Length | 114 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.88 |
| Shannon entropy | 0.51577 |
| G+C content | 0.56737 |
| Mean single sequence MFE | -38.45 |
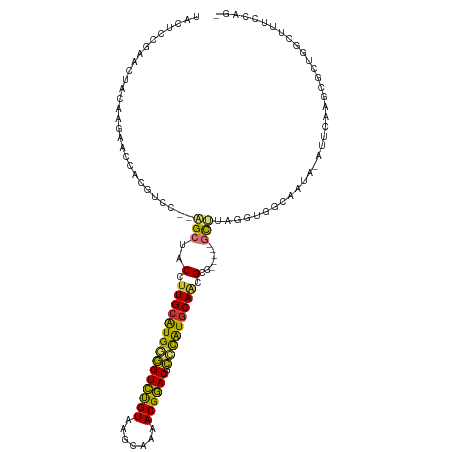
| Consensus MFE | -23.42 |
| Energy contribution | -22.19 |
| Covariance contribution | -1.23 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1867321 114 - 21146708 AUUUGUGCAGAGCAGCGCCGGCAGAACGAUGUGGUACGAUACUCCGAACUACAAGAACCACGUCCAGCUUCCUUGCGUGGGGUCUGUAAGCAAAACGGACGCCCAUGCAACGCG---- .....(((...)))(((..((.((...((((((((.(.................).))))))))...)).))(((((((((((((((.......)))))).)))))))))))).---- ( -40.63, z-score = -1.74, R) >droEre2.scaffold_4929 6174390 104 + 26641161 ----------UGCAGAGUCAGCAGAGCGACGUGGUACGAUAUUCCGAACUACAAGAACCACAUCCAGCUUCCUUGCAUGGGGUCUGUAAGCAAAACGGACGCCUGUGCAACGCG---- ----------(((.......)))..(((..(((((.((......)).)))))....................(((((((((((((((.......)))))).)))))))))))).---- ( -32.50, z-score = -1.18, R) >droYak2.chr2L 1681838 109 + 22324452 -----UGCAGAGCAGAGCCAGCAGAGCGACGUGGUACGUUACUCCGAACUACAAGAACCACGCCCAGCUACCUUGCAUGGGGUCUGUAAGCAAAACGGACGCCUAUUCAACGCG---- -----(((.(.((...))).)))..(((..(((((.((......)).)))))..(((.....((((((......)).))))((((((.......)))))).....)))..))).---- ( -29.00, z-score = 0.26, R) >droSec1.super_26 512148 109 - 826190 -----UGCAGAGCAGCGCCAGCAGAGCGAUGUGGUACGAUACUCCGAACUACAAGAUCCACGUCCAGCUACCUUGCAUGGGGUCUGUAAGCAAAACGGACGCCUAUGCAACGCG---- -----.((..(((.((....))...(.(((((((.......................)))))))).)))...(((((((((((((((.......)))))).))))))))).)).---- ( -36.60, z-score = -1.40, R) >droSim1.chr2R 765999 109 - 19596830 -----UGCAGAGCAGCGCCAGCAGAGCGAUGUGGUACGAUACUCCGAACUACAAGAACCACGUCCAGCUACCUUGCAUGGGGUCUGUAAGCAAAACGGACGCCUAUGCACCGCG---- -----.((..(((.((....))...(.((((((((.(.................).))))))))).)))....((((((((((((((.......)))))).))))))))..)).---- ( -37.93, z-score = -1.89, R) >droPer1.super_4 53600 109 + 7162766 -----UGAGGAGCCGAGCCGGCAGAGCGACGUGGUACGUUACACUAAUCAACUG----CGUUCUGGGCAGCCUUGCAUGAGGUCUGUAAGUGCCACGGACCCGCGUGCAAGGCGUAGG -----.......((..(((..(((((((...((((...........))))....----)))))))))).((((((((((.(((((((.......)))))))..))))))))))...)) ( -44.80, z-score = -1.49, R) >dp4.chr3 8072752 109 + 19779522 -----UGAGGAGCCGAGCCGGCAGAGCGACGUGGUACGUUACACUAAUCAACUG----CGUUCUGGGCAGCCUUGCAUGAGGUCUGUAAGUGCCACGGACCCGCGUGCAAGGCGUAGG -----.......((..(((..(((((((...((((...........))))....----)))))))))).((((((((((.(((((((.......)))))))..))))))))))...)) ( -44.80, z-score = -1.49, R) >droAna3.scaffold_13266 962895 100 + 19884421 -----AGCAACUAAGGGCCAUCAGAGCGAAGUGGUACGAAA--CUGAAAAA-------GGCGCCCGGCAGCCUUGCAUGUUGUUCGUAACAGAUACGAACCGACAUGCAGGGCG---- -----.((......(((((.((((..((........))...--))))....-------.).)))).)).(((((((((((((((((((.....)))))).))))))))))))).---- ( -41.30, z-score = -4.24, R) >consensus _____UGCAGAGCAGAGCCAGCAGAGCGACGUGGUACGAUACUCCGAACUACAAGAACCACGCCCAGCUACCUUGCAUGGGGUCUGUAAGCAAAACGGACGCCCAUGCAACGCG____ ..............(..(((.(........))))..).............................((....(((((((((((((((.......)))))).))))))))).))..... (-23.42 = -22.19 + -1.23)

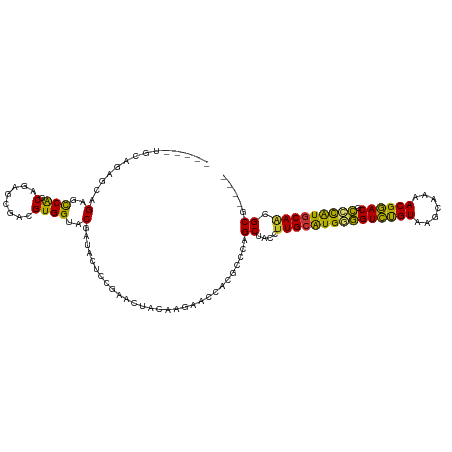
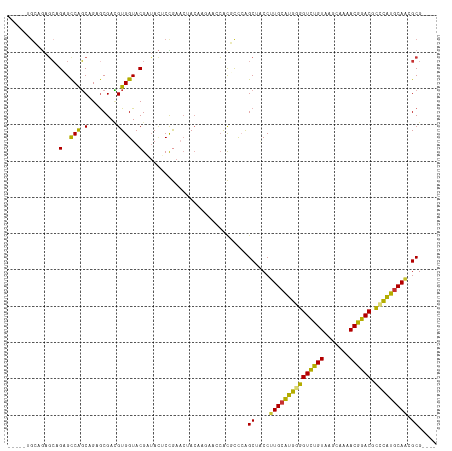
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:46 2011