
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,820,025 – 1,820,124 |
| Length | 99 |
| Max. P | 0.703524 |

| Location | 1,820,025 – 1,820,124 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.33 |
| Shannon entropy | 0.33805 |
| G+C content | 0.39224 |
| Mean single sequence MFE | -25.21 |
| Consensus MFE | -18.12 |
| Energy contribution | -18.23 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.703524 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 1820025 99 - 21146708 ACAAGAGAUUUAUGCGCUUGCCAAGCUGCAAGCAAGCAAACGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUGCAUUUCUCCA--GUCCUCUUUUU-------- ..((((((....(((((((((......))))))..)))...((((.((((.....))))......((((......)))).........--)))))))))).-------- ( -25.70, z-score = -2.00, R) >droSim1.chr2R_random 477226 99 - 2996586 ACAAGAGAUUUAUGCGCUUGCCAAGCUGCAAGCAAGCAAACGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUGCAUUUCUCCA--GUCCUCUUUUU-------- ..((((((....(((((((((......))))))..)))...((((.((((.....))))......((((......)))).........--)))))))))).-------- ( -25.70, z-score = -2.00, R) >droSec1.super_26 461013 99 - 826190 ACAAGAGAUUUAUGCGCUUGCCAAGCUGCAAGCAAGCAAACGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUGCAUUUCUCCA--GUCCUCUUUUU-------- ..((((((....(((((((((......))))))..)))...((((.((((.....))))......((((......)))).........--)))))))))).-------- ( -25.70, z-score = -2.00, R) >droYak2.chr2L 1598557 99 + 22324452 ACAAGAGAUUUAUGUGCUUGCCAAGCUGCAAGCAAGCAAACGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUGCAUUUCUCCA--GUCCUCUUUUU-------- ..((((((......(((((((......))))))).......((((.((((.....))))......((((......)))).........--)))))))))).-------- ( -25.00, z-score = -1.73, R) >droEre2.scaffold_4929 6104754 99 + 26641161 ACAAGAGAUUUAUGCGCUUGCCAAGCUGCAAGCAAGCAAACGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUGCAUUUCUCCA--GUCCUCUUUUU-------- ..((((((....(((((((((......))))))..)))...((((.((((.....))))......((((......)))).........--)))))))))).-------- ( -25.70, z-score = -2.00, R) >droAna3.scaffold_13266 921665 100 + 19884421 ACAGGAGAUUUAUGCGCUUGCCAAGCUACAAGCAAGAAAACGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUGCAUUUCUCCA-GUCCCUCCAUUU-------- ...(((((...(((((((((........))))).......((((..((((.....))))..))))............)))).))))).-............-------- ( -26.40, z-score = -2.81, R) >dp4.chr3 8048706 108 + 19779522 ACACAAGAUUUAUGCGCUUGCCAGGCUGCAAGCAAGCAAACGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUCCAUUUCUCCA-GUCGCAUUCCCACUCUGUCU ............(((((((((......))))))..)))...((((((.........(((...((((...................)))-)..)))........)))))) ( -23.44, z-score = -0.94, R) >droPer1.super_4 29577 108 + 7162766 ACACAAGAUUUAUGCGCUUGCCAGGCUGCAAGCAAGCAAACGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUCCAUUUCUCCA-GUCGCAUUCCCACUCUGUCU ............(((((((((......))))))..)))...((((((.........(((...((((...................)))-)..)))........)))))) ( -23.44, z-score = -0.94, R) >droWil1.scaffold_180700 2030950 107 - 6630534 ACAAAAGAUUUAUGCGCUUGCCAAGCUGCAAGCAAGCAAACGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUGCAUUUUCUCUAAUCGCCAUUCUUUGUUUU-- ...(((((.....((((((((......))))))..(((((((((..((((.....))))..)))).........))))).............))...))))).....-- ( -23.00, z-score = -0.43, R) >droVir3.scaffold_12823 1666112 107 + 2474545 ACUUUAGAUUUAUGCGCUUGCCAAGCUGCAAGCAAGCAAGCGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUGCAUUGCUCUA-GUCGCUCGGGUUUUUUCCG- .............((((((((......)))))).(((((.((((..((((.....))))..))))((((......)))).)))))...-...)).((((.....))))- ( -27.30, z-score = -0.29, R) >droMoj3.scaffold_6496 3161838 98 - 26866924 -UUUUAGAUUUAUGCGCUUGCCAAGCUGCAAGCAAGCAAACGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUCCAUUCCAGCU--CUGUUUUCCCU-------- -...........(((((((((......))))))..)))((((((..((((.....))))...((((................)))).)--)))))......-------- ( -21.29, z-score = -0.88, R) >droGri2.scaffold_15245 8870380 107 - 18325388 ACGGGAGAUUUAUGCGCUUGCCAAGCUGCAAGCAAGAAAACGGACAGCAAUUAAUUUGCAUUCUAGUAAUUAAUUUUGCAUUGCUGUA-GUCGCCCGAUUUUUUUCUU- .((((.((((...((((((((......))))))........(((..((((.....))))..))).((((......))))...))...)-))).))))...........- ( -29.80, z-score = -1.80, R) >consensus ACAAGAGAUUUAUGCGCUUGCCAAGCUGCAAGCAAGCAAACGGACAGCAAUUAAUUUGCAUUCUGGUAAUUAAUUUUGCAUUUCUCCA__UCCCUCUUUUU________ ....((((((..(((((((((......)))))).......((((..((((.....))))..)))))))...))))))................................ (-18.12 = -18.23 + 0.11)
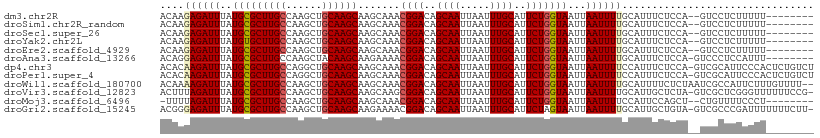

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:42 2011