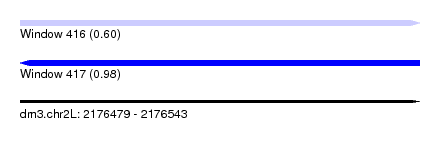
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,176,479 – 2,176,543 |
| Length | 64 |
| Max. P | 0.982417 |

| Location | 2,176,479 – 2,176,543 |
|---|---|
| Length | 64 |
| Sequences | 3 |
| Columns | 71 |
| Reading direction | forward |
| Mean pairwise identity | 54.37 |
| Shannon entropy | 0.61373 |
| G+C content | 0.29571 |
| Mean single sequence MFE | -13.87 |
| Consensus MFE | -3.23 |
| Energy contribution | -4.79 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
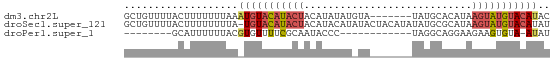
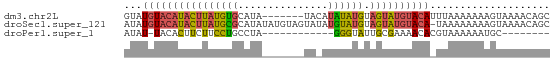
>dm3.chr2L 2176479 64 + 23011544 GCUGUUUUACUUUUUUUAAAUGUACAUACUACAUAUAUGUA-------UAUGCACAUAAGUAUGUACAUAC ...................(((((((((((.....(((((.-------.....)))))))))))))))).. ( -14.80, z-score = -2.28, R) >droSec1.super_121 48961 70 + 59987 GCUGUUUUACUUUUUUUUA-UGUACAUACUACAUACAUAUACUACAUAUAUGCGCAUAAGUAUGUACAUAU .................((-((((((((((.....((((((.....))))))......)))))))))))). ( -16.90, z-score = -3.49, R) >droPer1.super_1 4540392 50 - 10282868 --------GCAUUUUUUACGUGUUUUCGCAAUACCC------------UAGGCAGGAAGAAGUGUA-AUAU --------(((((((((.(.(((((...........------------.))))).)))))))))).-.... ( -9.90, z-score = -1.28, R) >consensus GCUGUUUUACUUUUUUUAA_UGUACAUACUACAUACAU_UA_______UAUGCACAUAAGUAUGUACAUAU ...................(((((((((((............................))))))))))).. ( -3.23 = -4.79 + 1.56)
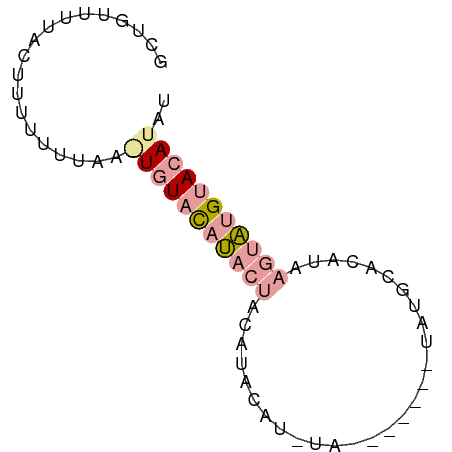
| Location | 2,176,479 – 2,176,543 |
|---|---|
| Length | 64 |
| Sequences | 3 |
| Columns | 71 |
| Reading direction | reverse |
| Mean pairwise identity | 54.37 |
| Shannon entropy | 0.61373 |
| G+C content | 0.29571 |
| Mean single sequence MFE | -14.23 |
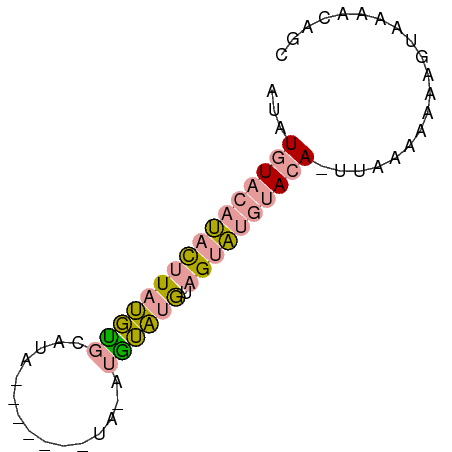
| Consensus MFE | -6.94 |
| Energy contribution | -7.39 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2176479 64 - 23011544 GUAUGUACAUACUUAUGUGCAUA-------UACAUAUAUGUAGUAUGUACAUUUAAAAAAAGUAAAACAGC ..(((((((((((((((((....-------..))))))...)))))))))))................... ( -15.90, z-score = -2.13, R) >droSec1.super_121 48961 70 - 59987 AUAUGUACAUACUUAUGCGCAUAUAUGUAGUAUAUGUAUGUAGUAUGUACA-UAAAAAAAAGUAAAACAGC .((((((((((((.((..((((((((...))))))))..))))))))))))-))................. ( -21.90, z-score = -3.71, R) >droPer1.super_1 4540392 50 + 10282868 AUAU-UACACUUCUUCCUGCCUA------------GGGUAUUGCGAAAACACGUAAAAAAUGC-------- ....-.........((((....)------------)))..(((((......))))).......-------- ( -4.90, z-score = 0.38, R) >consensus AUAUGUACAUACUUAUGUGCAUA_______UA_AUGUAUGUAGUAUGUACA_UUAAAAAAAGUAAAACAGC ...((((((((((((((((...............)))))).)))))))))).................... ( -6.94 = -7.39 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:26 2011