| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,782,956 – 1,783,056 |
| Length | 100 |
| Max. P | 0.543088 |

| Location | 1,782,956 – 1,783,056 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.58 |
| Shannon entropy | 0.38291 |
| G+C content | 0.49661 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.91 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.543088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1782956 100 + 21146708 --------CAUGAGCCUGGUUAGUAAAGACUUAAGCUGACAGCCGCCACCGAGCGAAACGCAGAACAGUUUGAGUUACUUUCUACCCAGCAUUUCA---UCCGCUCAGCCG --------..(((((((((.(((.((((((((((((((...(((((......)))....))....))))))))))..))))))).)))).......---...))))).... ( -28.00, z-score = -2.62, R) >droSim1.chr2R 691853 100 + 19596830 --------CAUGAGCCUGGUUAGUAAAGACUUAAGCUGACAGCCGCCACCGAGCGAAACGCAGAACAGUUUGAGUUACUUUCUACCCAGCAUUUCA---UCCGCUCAGCCG --------..(((((((((.(((.((((((((((((((...(((((......)))....))....))))))))))..))))))).)))).......---...))))).... ( -28.00, z-score = -2.62, R) >droSec1.super_26 432542 100 + 826190 --------CAUGAGCCUGGUUAGUAAAGACUUAAGCUGACAGCCGCCACCGAGCGAAACGCAGAACAGUUUGAGUUACUUUCUAUCCAGCAUUUCA---UCCGCUCAGCCG --------..(((((((((.(((.((((((((((((((...(((((......)))....))....))))))))))..))))))).)))).......---...))))).... ( -28.10, z-score = -2.49, R) >droYak2.chr2L 1566217 100 - 22324452 --------CAUGAGCCUGGUUAGUAAAGACUUAAGCUGACAGCAGCCACCGAGCGAAACGCAGAACAGUUUGAGUUACUUUCUACACAGCAUUUUA---UCCGCUCAGCCG --------..((((((((..(((.((((((((((((((...((.((......)).....))....))))))))))..)))))))..))).......---...))))).... ( -23.30, z-score = -0.91, R) >droEre2.scaffold_4929 6075485 100 - 26641161 --------CAUGAGCCUGGUUAGUAAAGACUUAAGCUGACAGCCGCCACCGAGCGAAGCGCAGAACAGUUUGAGUUACUUUCUACCCAGAAUUUUA---UCCGCUCAGCCG --------..(((((((((.(((.((((((((((((((...(((((......)))..))......))))))))))..))))))).)))).......---...))))).... ( -28.70, z-score = -2.76, R) >droAna3.scaffold_13266 881116 92 - 19884421 --------CCUCAGCCUGGUUAGUGGCAACUUAAGCUGACAGCAAC------GCAGAGCGGCGAGCAGUUUGAGUUACUUUCUAC-CAGCAUUCU----UUGGCUAAGCCC --------.....((.((((....((.(((((((((((...((..(------((...)))))...))))))))))).))....))-)))).....----..(((...))). ( -28.30, z-score = -0.47, R) >droWil1.scaffold_180700 4549807 99 - 6630534 GCUUCAGUUUUCAGCUUGGCCAAC--UGACUUAAGCUGACAGC----------CUAGCAGCCGCACAGUUUGAGUUACUUUCUAAAUAGCAUUUUACGUUUGCCUCUGCUG ...........((((..(((.(((--((((((((((((.(.((----------......)).)..)))))))))))).....((((......)))).))).)))...)))) ( -25.00, z-score = -0.61, R) >consensus ________CAUGAGCCUGGUUAGUAAAGACUUAAGCUGACAGCCGCCACCGAGCGAAACGCAGAACAGUUUGAGUUACUUUCUACCCAGCAUUUUA___UCCGCUCAGCCG ..........((((((((..(((....(((((((((((...((................))....))))))))))).....)))..))).............))))).... (-14.28 = -14.91 + 0.64)

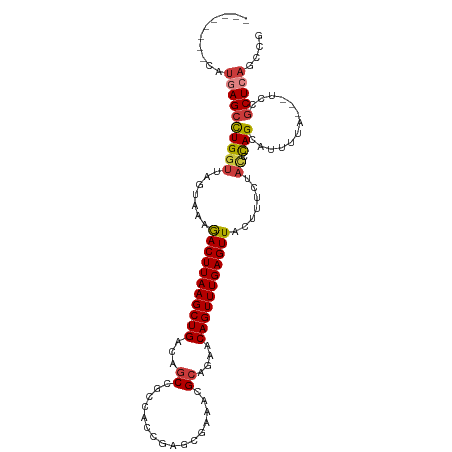
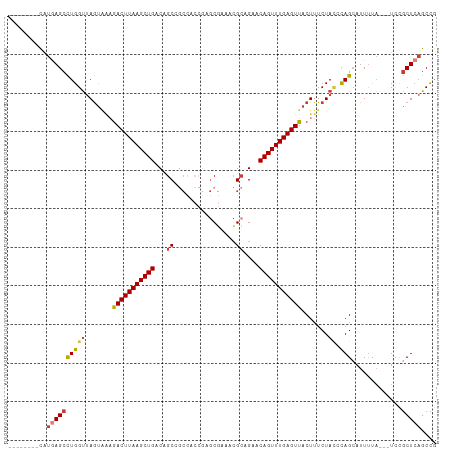
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:41 2011