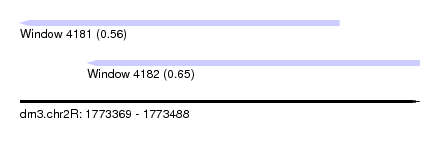
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,773,369 – 1,773,488 |
| Length | 119 |
| Max. P | 0.645952 |

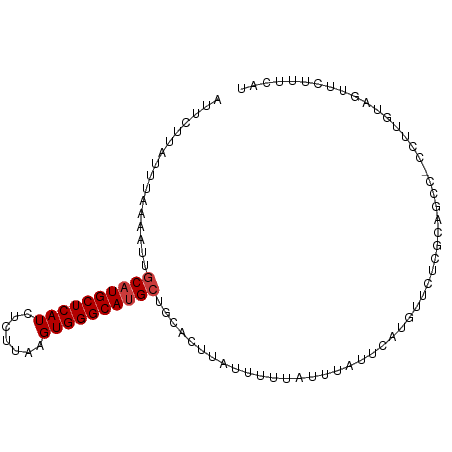
| Location | 1,773,369 – 1,773,464 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 88.76 |
| Shannon entropy | 0.22853 |
| G+C content | 0.33999 |
| Mean single sequence MFE | -15.55 |
| Consensus MFE | -13.23 |
| Energy contribution | -13.51 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.564549 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 1773369 95 - 21146708 AUUCUUAUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCAUGCUGCACUUAUUUUUAUUUAUUCAUGUUCUCACAGCC-CCUUGUAGUUCUUUCAU ................((((((((((.......)))))))))).......................(..((.((((..-..))))))..)...... ( -15.50, z-score = -1.51, R) >droSim1.chr2R 683099 95 - 19596830 AUUCUUAUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCAUGCUGCACUUAUUUUUAUUUAUUCAUGUUCUCGCCGCC-CCUUGCAGUUCUUUCAU ................((((((((((.......)))))))))).....................(((.....((.((.-....)).)).....))) ( -15.90, z-score = -1.37, R) >droSec1.super_26 423613 95 - 826190 AUUCUUAUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCAUGCUGCACUUAUUUUUAUUUAUUCAUGUUCUCGCCGCC-CCUUGUAGUUCUUUCAU ................((((((((((.......)))))))))).....................(((.....((..(.-....)..)).....))) ( -14.80, z-score = -1.28, R) >droYak2.chr2L 1557209 95 + 22324452 AUUCUUAUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCAUGGUGCACUUAUUUUUAUUUAUUCAUGUUCUCGCAGCC-CCUUGUAGUUCUUUCAU ............((((((((((((((.......))))))))(((((..........................))).))-....))))))....... ( -13.87, z-score = -0.32, R) >droEre2.scaffold_4929 6066744 95 + 26641161 AUUCUUAUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCAUGCUGCACUUAUUUUUAUUUAUUCAUGCUCUCGCAGCC-CCUUGUAGUUCUUUCAU ............((((((((((((((.......))))))))(((((..........................))))).-....))))))....... ( -17.67, z-score = -1.63, R) >droAna3.scaffold_13266 19013755 94 - 19884421 AUUCUUGUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCA-GCUGCAUUUAAUUUUAUUUAUUCAUGUUUACCCAGCC-UCGCUACGUUCUUUCAA .....((..(((((((((((((((((.......)))))))-..))))....))))))..)).............(((.-..)))............ ( -12.20, z-score = 0.15, R) >droVir3.scaffold_12823 1596252 96 + 2474545 AUUCUUAUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCAUGCUGCAUUUAAUUUUAUUUAUUCAUGUUUCUUUCUCUAUACUAUAGAGUAUAUAU .....((..(((((((((((((((((.......)))))))))).......)))))))..)).............((((((...))))))....... ( -18.91, z-score = -2.70, R) >consensus AUUCUUAUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCAUGCUGCACUUAUUUUUAUUUAUUCAUGUUCUCGCAGCC_CCUUGUAGUUCUUUCAU ................((((((((((.......))))))))))..................................................... (-13.23 = -13.51 + 0.29)


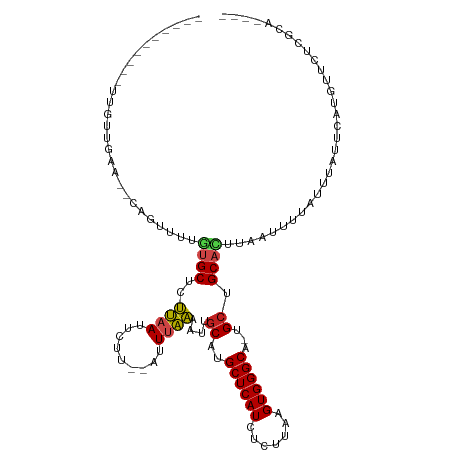
| Location | 1,773,389 – 1,773,488 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.57 |
| Shannon entropy | 0.44296 |
| G+C content | 0.33250 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -8.97 |
| Energy contribution | -8.77 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.645952 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 1773389 99 - 21146708 ----------UUGUUGAA--CAGUUUUGUGCUCUUAAUUCUU--AUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCA-UGCUGCACUUAUUUUUAUUUAUUCAUGUUCUCACA---- ----------.(((.(((--((.....((((..((((.....--..))))....((((((((((.......)))))))-))).)))).................)))))..)))---- ( -21.55, z-score = -2.76, R) >droSim1.chr2R 683119 99 - 19596830 ----------UUGUUGAA--CAGUUUUGUGCUCUUAAUUCUU--AUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCA-UGCUGCACUUAUUUUUAUUUAUUCAUGUUCUCGCC---- ----------.....(((--((.....((((..((((.....--..))))....((((((((((.......)))))))-))).)))).................))))).....---- ( -21.25, z-score = -2.63, R) >droSec1.super_26 423633 99 - 826190 ----------UUGUUGAA--CAGUUUUGUGCUCUUAAUUCUU--AUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCA-UGCUGCACUUAUUUUUAUUUAUUCAUGUUCUCGCC---- ----------.....(((--((.....((((..((((.....--..))))....((((((((((.......)))))))-))).)))).................))))).....---- ( -21.25, z-score = -2.63, R) >droYak2.chr2L 1557229 99 + 22324452 ----------UUGUUGAA--AAGUUUUGUGCUCUUAAUUCUU--AUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCA-UGGUGCACUUAUUUUUAUUUAUUCAUGUUCUCGCA---- ----------......((--((((...((((.(((((.....--..)))).....(((((((((.......)))))))-))).))))..))))))...................---- ( -18.60, z-score = -1.48, R) >droEre2.scaffold_4929 6066764 99 + 26641161 ----------UUGUUGAA--CAGUUUUGUGCUCUUAAUUCUU--AUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCA-UGCUGCACUUAUUUUUAUUUAUUCAUGCUCUCGCA---- ----------........--.......((((..((((.....--..))))....((((((((((.......)))))))-))).)))).................(((....)))---- ( -20.00, z-score = -1.60, R) >dp4.chr3 8013453 85 + 19779522 ---------------------------CCGCUCGACAGUCUUGGGCUUUUAGUUGCAUGCUCAUCUCUUGAGUGGGCU-GGCUGCAUUUAAUUUUAUUUAUUCAUGUUCUCCC----- ---------------------------((((((((.((...(((((............))))).)).))))))))...-((..((((.(((......)))...))))...)).----- ( -20.50, z-score = -1.02, R) >droPer1.super_5770 475 83 + 1806 ---------------------------CCGCUCGACAGUCUUGGGCUUUUAGUUGCAUGCUCAUCUCUUAAGUGGGCU-GGCUGCAUU--AAUUUAUUUAUUCAUGUUCUCCC----- ---------------------------(((((..(.((...(((((............))))).)).)..)))))...-((..((((.--(((......))).))))...)).----- ( -17.10, z-score = -0.16, R) >droAna3.scaffold_13266 19013775 90 - 19884421 --------------------CAGUUCUAUGCUUUUAAUUCUU--GUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCA--GCUGCAUUUAAUUUUAUUUAUUCAUGUUUACCCA---- --------------------.((((..((((((((((.....--..))))))..((.(((((((.......)))))))--)).))))..)))).....................---- ( -14.50, z-score = -0.75, R) >droWil1.scaffold_180700 4538916 99 + 6630534 ----------UCGUUUAA--CAGUUUUGUGCUUUUAAUUCUU--AUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCA-UGCUGCAUUUAAUUUUAUUUAUUCAUGUUUUUUUC---- ----------........--.((((..((((((((((.....--..))))))..((((((((((.......)))))))-))).))))..)))).....................---- ( -20.20, z-score = -3.19, R) >droGri2.scaffold_15245 8815295 113 - 18325388 CUCACUAUCUUUGCUUAA--CAGUUUGGUGCUUUUAAUUCUU--AUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCU-UGCUGCAUUUAAUUUUAUUUAUUCAUGUUUCUUUUUAAA ..................--.((((..((((((((((.....--..))))))..(((.((((((.......)))))).-))).))))..))))......................... ( -16.10, z-score = -0.63, R) >droMoj3.scaffold_6496 3076805 110 - 26866924 ------CUGUUCAUCAAGUUUGUUUCUGUGCUUUUAAUUCUU--AUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCAAUGCUGCACUUAAUUUUAUUUAUUCAUGUUUCCUUUCUAU ------.........((((..(((...((((((((((.....--..))))))..((((((((((.......)))))).)))).))))..)))...))))................... ( -17.30, z-score = -1.68, R) >droVir3.scaffold_12823 1596270 102 + 2474545 ---------CUCGUCAA---CAGUUUUGUGCUUUUAAUUCUU--AUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCA-UGCUGCAUUUAAUUUUAUUUAUUCAUGUUUCUUUCUCU- ---------........---.((((..((((((((((.....--..))))))..((((((((((.......)))))))-))).))))..))))........................- ( -20.20, z-score = -3.59, R) >consensus __________UUGUUGAA__CAGUUUUGUGCUCUUAAUUCUU__AUUUAAAAUUGCAUGCUCAUCUCUUAAGUGGGCA_UGCUGCACUUAAUUUUAUUUAUUCAUGUUCUCGCA____ ...........................((((..((((.........))))....((..((((((.......))))))...)).))))............................... ( -8.97 = -8.77 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:39 2011