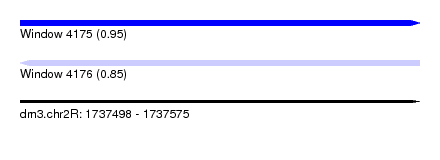
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,737,498 – 1,737,575 |
| Length | 77 |
| Max. P | 0.946867 |

| Location | 1,737,498 – 1,737,575 |
|---|---|
| Length | 77 |
| Sequences | 11 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 83.45 |
| Shannon entropy | 0.35937 |
| G+C content | 0.40286 |
| Mean single sequence MFE | -14.38 |
| Consensus MFE | -10.12 |
| Energy contribution | -9.86 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946867 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
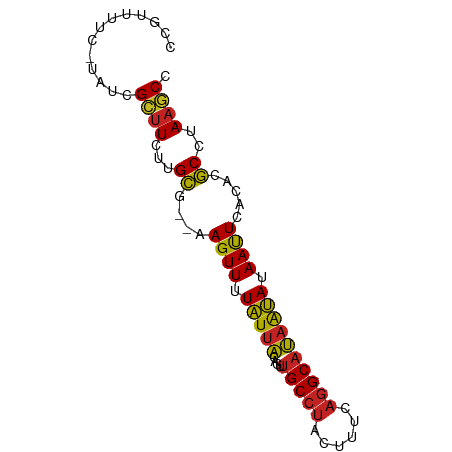
>dm3.chr2R 1737498 77 + 21146708 CCGUUUCC-UAUCGCUUCUUGCG--AAGUUUUAUUACUGUUGCCUACUUCCAGGCAUAAUAUAAUUCACACGCCUAAGCC ........-....((((...(((--..(..((((......(((((......)))))....))))..)...)))..)))). ( -13.60, z-score = -1.86, R) >droSim1.chr2R 651130 77 + 19596830 CCGUUUCC-UAUCGCUUCUUGCG--AAGUUUUAUUACUGUUGCCUACUUCCAGGCAUAAUAUAAUUCACACGCCUAAGCC ........-....((((...(((--..(..((((......(((((......)))))....))))..)...)))..)))). ( -13.60, z-score = -1.86, R) >droSec1.super_26 393908 77 + 826190 CCGUUUCC-UAUCGCUUCUUGCG--AAGUUUUAUUACUGUUGCCUACUUCCAGGCAUAGUAUAAUUCACACGCCUAAGCC ........-....((((...(((--..(..((((.(((((.((((......)))))))))))))..)...)))..)))). ( -17.50, z-score = -2.96, R) >droYak2.chr2L 1524180 77 - 22324452 CCGUUUAC-UAUCGCCUCUUGCG--AAGUUUUAUUACAGUUGCCUACUUCCAGGCAUAAUAUAACUCACACGCCUAAGCC ..(((((.-..((((.....)))--)((((.(((((....(((((......)))))))))).))))........))))). ( -15.30, z-score = -2.34, R) >droEre2.scaffold_4929 6035126 77 - 26641161 CCGCUUGC-CUUCGCUUCUGGCG--AAGUUUUAUUACACUUGCCUACUUCCAGGCAUAAUAUAAUUCACACGCCUAAGCC ..((((((-((((((.....)))--)))............(((((......)))))...............))..)))). ( -18.50, z-score = -2.82, R) >droAna3.scaffold_13266 841957 77 - 19884421 -CCUUUUC-UAUCGCUUUUUGCGC-AAGUUUUAUUACAGUUGCCUACUUUCAGGCAUAAUAUAAUUCACACGCCUAAGCC -.......-....((((...(((.-..(..((((......(((((......)))))....))))..)...)))..)))). ( -14.60, z-score = -2.51, R) >dp4.chr3 7990006 78 - 19779522 CCUUUUUC-UAUCGCUUCUUGCGC-AAGUUUUAUUACUGUUGCCUACUUUCAGGCAUAAUAUAAUUCACACGCCUAAGCC ........-....((((...(((.-..(..((((......(((((......)))))....))))..)...)))..)))). ( -13.90, z-score = -2.10, R) >droPer1.super_7823 202 77 + 9810 -CUUUUUUUUUUCGUUUUUGGGC--GAGUUUUUUUUUUUGGGCGCUUUUUUUGGCGAAAAAUAAUUUAAACCCCCAAACC -...............((((((.--(.((((.((.((((...((((......)))).)))).))...))))))))))).. ( -15.20, z-score = -0.57, R) >droWil1.scaffold_180700 4496754 78 - 6630534 CCCAUUUC-UAUUGCUUUUUGCUC-AAGUUUUAUUACAGUUGCCUACUUUCAGGCAUAAUAUAAUUCACACGCCUAAGCC ........-....((((...((..-..(..((((......(((((......)))))....))))..)....))..)))). ( -11.60, z-score = -1.54, R) >droVir3.scaffold_12823 1558625 80 - 2474545 CCAUUUCUAUUUUGCUUUUUGCUCCAAGUUAUAUUACAGUUGCCUAUUUUCAGGCAUUAUAUAAUUCACACGCCUAAACC .............((.....))....((((((((......(((((......)))))..)))))))).............. ( -10.10, z-score = -1.42, R) >droGri2.scaffold_15245 8784410 77 + 18325388 -CCAUUUC-CAUUGCUUUUUGCGC-AAGUUUUAUUACAGUUGCCUACUUUCAGGCAUAAUAUAAUUCACACGCCUAAGCC -.......-....((((...(((.-..(..((((......(((((......)))))....))))..)...)))..)))). ( -14.30, z-score = -2.31, R) >consensus CCGUUUUC_UAUCGCUUCUUGCG__AAGUUUUAUUACAGUUGCCUACUUUCAGGCAUAAUAUAAUUCACACGCCUAAGCC .............((((...((....((((.(((((....(((((......)))))))))).)))).....))..)))). (-10.12 = -9.86 + -0.26)

| Location | 1,737,498 – 1,737,575 |
|---|---|
| Length | 77 |
| Sequences | 11 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 83.45 |
| Shannon entropy | 0.35937 |
| G+C content | 0.40286 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -12.04 |
| Energy contribution | -12.05 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854381 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 1737498 77 - 21146708 GGCUUAGGCGUGUGAAUUAUAUUAUGCCUGGAAGUAGGCAACAGUAAUAAAACUU--CGCAAGAAGCGAUA-GGAAACGG .((((..(((.((..(((((....((((((....))))))...)))))...))..--)))...))))....-(....).. ( -20.40, z-score = -2.43, R) >droSim1.chr2R 651130 77 - 19596830 GGCUUAGGCGUGUGAAUUAUAUUAUGCCUGGAAGUAGGCAACAGUAAUAAAACUU--CGCAAGAAGCGAUA-GGAAACGG .((((..(((.((..(((((....((((((....))))))...)))))...))..--)))...))))....-(....).. ( -20.40, z-score = -2.43, R) >droSec1.super_26 393908 77 - 826190 GGCUUAGGCGUGUGAAUUAUACUAUGCCUGGAAGUAGGCAACAGUAAUAAAACUU--CGCAAGAAGCGAUA-GGAAACGG .((((..(((.((......((((.((((((....))))))..)))).....))..--)))...))))....-(....).. ( -20.90, z-score = -2.39, R) >droYak2.chr2L 1524180 77 + 22324452 GGCUUAGGCGUGUGAGUUAUAUUAUGCCUGGAAGUAGGCAACUGUAAUAAAACUU--CGCAAGAGGCGAUA-GUAAACGG .((((..(((...(((((.(((((((((((....))))))....))))).)))))--)))...))))....-........ ( -20.50, z-score = -1.53, R) >droEre2.scaffold_4929 6035126 77 + 26641161 GGCUUAGGCGUGUGAAUUAUAUUAUGCCUGGAAGUAGGCAAGUGUAAUAAAACUU--CGCCAGAAGCGAAG-GCAAGCGG .((((.((((.((..((((((((.((((((....))))))))))))))...))..--))))....((....-)))))).. ( -28.60, z-score = -4.00, R) >droAna3.scaffold_13266 841957 77 + 19884421 GGCUUAGGCGUGUGAAUUAUAUUAUGCCUGAAAGUAGGCAACUGUAAUAAAACUU-GCGCAAAAAGCGAUA-GAAAAGG- .((((..((((((..((((((...((((((....))))))..))))))...))..-))))...))))....-.......- ( -19.40, z-score = -2.15, R) >dp4.chr3 7990006 78 + 19779522 GGCUUAGGCGUGUGAAUUAUAUUAUGCCUGAAAGUAGGCAACAGUAAUAAAACUU-GCGCAAGAAGCGAUA-GAAAAAGG .((((..((((((..(((((....((((((....))))))...)))))...))..-))))...))))....-........ ( -19.10, z-score = -2.10, R) >droPer1.super_7823 202 77 - 9810 GGUUUGGGGGUUUAAAUUAUUUUUCGCCAAAAAAAGCGCCCAAAAAAAAAAACUC--GCCCAAAAACGAAAAAAAAAAG- ..(((((((((((...........(((........)))...........)))).)--.))))))...............- ( -11.15, z-score = 0.25, R) >droWil1.scaffold_180700 4496754 78 + 6630534 GGCUUAGGCGUGUGAAUUAUAUUAUGCCUGAAAGUAGGCAACUGUAAUAAAACUU-GAGCAAAAAGCAAUA-GAAAUGGG .(((((((..(....((((((...((((((....))))))..)))))).)..)))-))))...........-........ ( -16.60, z-score = -1.32, R) >droVir3.scaffold_12823 1558625 80 + 2474545 GGUUUAGGCGUGUGAAUUAUAUAAUGCCUGAAAAUAGGCAACUGUAAUAUAACUUGGAGCAAAAAGCAAAAUAGAAAUGG ..(((((((((............))))))))).((((....)))).............((.....))............. ( -15.50, z-score = -1.64, R) >droGri2.scaffold_15245 8784410 77 - 18325388 GGCUUAGGCGUGUGAAUUAUAUUAUGCCUGAAAGUAGGCAACUGUAAUAAAACUU-GCGCAAAAAGCAAUG-GAAAUGG- .((((..((((((..((((((...((((((....))))))..))))))...))..-))))...))))....-.......- ( -19.40, z-score = -1.95, R) >consensus GGCUUAGGCGUGUGAAUUAUAUUAUGCCUGAAAGUAGGCAACUGUAAUAAAACUU__CGCAAGAAGCGAUA_GAAAACGG .((((..((..((..(((((....((((((....))))))...)))))...)).....))...))))............. (-12.04 = -12.05 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:34 2011