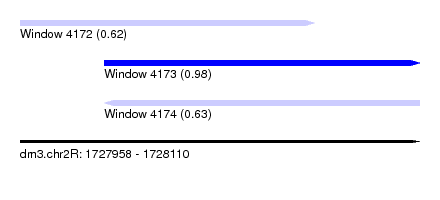
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,727,958 – 1,728,110 |
| Length | 152 |
| Max. P | 0.977075 |

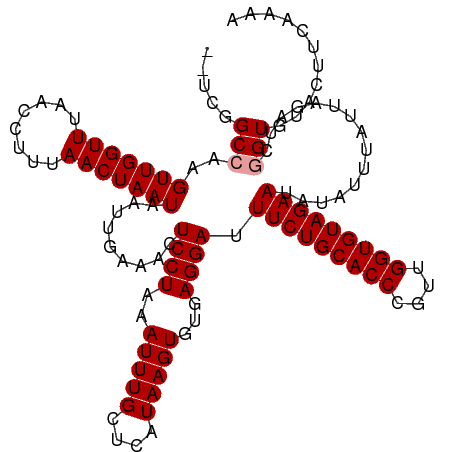
| Location | 1,727,958 – 1,728,070 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.83 |
| Shannon entropy | 0.15305 |
| G+C content | 0.34573 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.624954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1727958 112 + 21146708 --UCGGCCAAGUUGGUUUAACCUUUAACUAAUAA------CUCCUAAAUUUGCUCAUAAGUGUAAGGAUUUCUGCACCCAAUGGUGUAGAAUACAUUUAUUAAGUCGGUGCACUUAAAAA --..((....(((((((........)))))))..------..))............((((((((..((((((((((((....)))))))))............)))..)))))))).... ( -27.10, z-score = -2.26, R) >droSec1.super_26 384624 118 + 826190 --UCGGCCAAGUUGGUUUAACCUUUAACUAAUAAUUGAAACUCCUAAAUUUGCUCAUAAGUGUGAGGAUUUCUGCACCAGUUGGUGUAGAAUAUAUUUAUUAGAUCGGUGUACUUCAAAA --...(((.....((.....)).....(((((((.((....((((..(((((....)))))...)))).(((((((((....)))))))))..)).)))))))...)))........... ( -27.50, z-score = -1.72, R) >droSim1.chr2R 641792 120 + 19596830 UCACGGCCAAGUUGGUUUAACCUUUAACUAAUAAUUGAAACUCCUAAAUUUGCUCAUAAGUGUGAGGAUUUCUGCACCCGUUGGUGUAGAAUAUAUUUCUUAGAUCAGUGUACUUCAAAA .((((..(..(((((((........)))))))....((((.((((..(((((....)))))...)))).(((((((((....)))))))))....))))...)..).))).......... ( -24.70, z-score = -0.89, R) >consensus __UCGGCCAAGUUGGUUUAACCUUUAACUAAUAAUUGAAACUCCUAAAUUUGCUCAUAAGUGUGAGGAUUUCUGCACCCGUUGGUGUAGAAUAUAUUUAUUAGAUCGGUGUACUUCAAAA .....(((..(((((((........))))))).........((((..(((((....)))))...)))).(((((((((....)))))))))...............)))........... (-22.20 = -22.53 + 0.33)

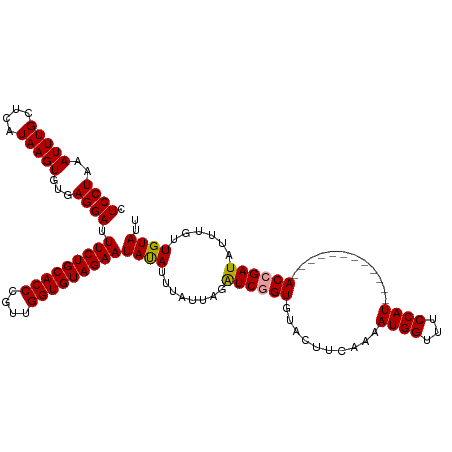
| Location | 1,727,990 – 1,728,110 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.78 |
| Shannon entropy | 0.17601 |
| G+C content | 0.34815 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -23.46 |
| Energy contribution | -23.36 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1727990 120 + 21146708 CUCCUAAAUUUGCUCAUAAGUGUAAGGAUUUCUGCACCCAAUGGUGUAGAAUACAUUUAUUAAGUCGGUGCACUUAAAAAUGGUUUCCAUCCCCAUAUAUGUACCGAUAUUUGUUGUAUU ..........(((..(((((((((.....(((((((((....))))))))))))))))))...(((((((((..((...((((...)))).....))..))))))))).......))).. ( -31.70, z-score = -3.34, R) >droSec1.super_26 384662 108 + 826190 CUCCUAAAUUUGCUCAUAAGUGUGAGGAUUUCUGCACCAGUUGGUGUAGAAUAUAUUUAUUAGAUCGGUGUACUUCAAAAUGGUUUCCAU------------ACCGAUAUUUGUUGUAUU ...((((.....(((((....)))))...(((((((((....)))))))))........))))(((((((((((.......)))....))------------))))))............ ( -28.10, z-score = -2.67, R) >droSim1.chr2R 641832 108 + 19596830 CUCCUAAAUUUGCUCAUAAGUGUGAGGAUUUCUGCACCCGUUGGUGUAGAAUAUAUUUCUUAGAUCAGUGUACUUCAAAAUGGUUUCCAU------------ACCGAUAUUUGUUGUAUU ...((((.....(((((....)))))...(((((((((....)))))))))........))))......((((..((((.((((......------------))))...))))..)))). ( -24.10, z-score = -1.71, R) >consensus CUCCUAAAUUUGCUCAUAAGUGUGAGGAUUUCUGCACCCGUUGGUGUAGAAUAUAUUUAUUAGAUCGGUGUACUUCAAAAUGGUUUCCAU____________ACCGAUAUUUGUUGUAUU .((((..(((((....)))))...)))).(((((((((....)))))))))((((........((((((..........((((...))))............))))))......)))).. (-23.46 = -23.36 + -0.11)

| Location | 1,727,990 – 1,728,110 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.78 |
| Shannon entropy | 0.17601 |
| G+C content | 0.34815 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -18.70 |
| Energy contribution | -17.82 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.632878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1727990 120 - 21146708 AAUACAACAAAUAUCGGUACAUAUAUGGGGAUGGAAACCAUUUUUAAGUGCACCGACUUAAUAAAUGUAUUCUACACCAUUGGGUGCAGAAAUCCUUACACUUAUGAGCAAAUUUAGGAG .............(((((.(((...(((((((((...))))))))).))).)))))((((.(((.((((((((.((((....)))).)))).....)))).)))))))............ ( -24.90, z-score = -1.28, R) >droSec1.super_26 384662 108 - 826190 AAUACAACAAAUAUCGGU------------AUGGAAACCAUUUUGAAGUACACCGAUCUAAUAAAUAUAUUCUACACCAACUGGUGCAGAAAUCCUCACACUUAUGAGCAAAUUUAGGAG ............((((((------------((((...))))..........))))))((..(((((...((((.((((....)))).))))...((((......))))...)))))..)) ( -18.60, z-score = -1.43, R) >droSim1.chr2R 641832 108 - 19596830 AAUACAACAAAUAUCGGU------------AUGGAAACCAUUUUGAAGUACACUGAUCUAAGAAAUAUAUUCUACACCAACGGGUGCAGAAAUCCUCACACUUAUGAGCAAAUUUAGGAG ..(((..((((....(((------------......)))..))))..)))......((((((.......((((.((((....)))).))))...((((......))))....)))))).. ( -18.10, z-score = -0.84, R) >consensus AAUACAACAAAUAUCGGU____________AUGGAAACCAUUUUGAAGUACACCGAUCUAAUAAAUAUAUUCUACACCAACGGGUGCAGAAAUCCUCACACUUAUGAGCAAAUUUAGGAG .............(((((............((((...))))..........)))))(((((........((((.((((....)))).))))...((((......)))).....))))).. (-18.70 = -17.82 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:32 2011