| Sequence ID | dm3.chr2R |
|---|---|
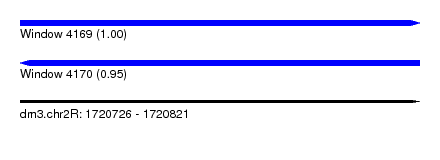
| Location | 1,720,726 – 1,720,821 |
| Length | 95 |
| Max. P | 0.996154 |

| Location | 1,720,726 – 1,720,821 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.85 |
| Shannon entropy | 0.35546 |
| G+C content | 0.49903 |
| Mean single sequence MFE | -37.93 |
| Consensus MFE | -27.35 |
| Energy contribution | -28.17 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
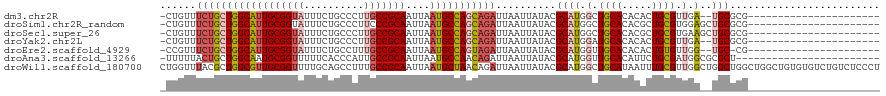
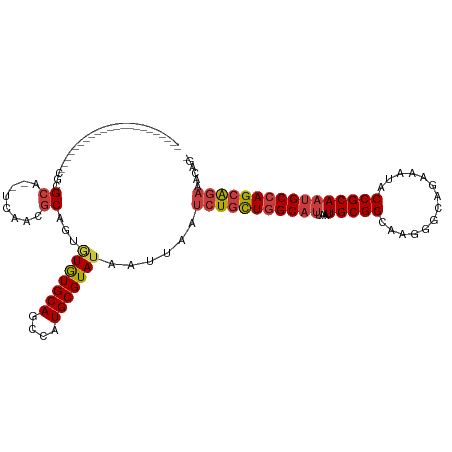
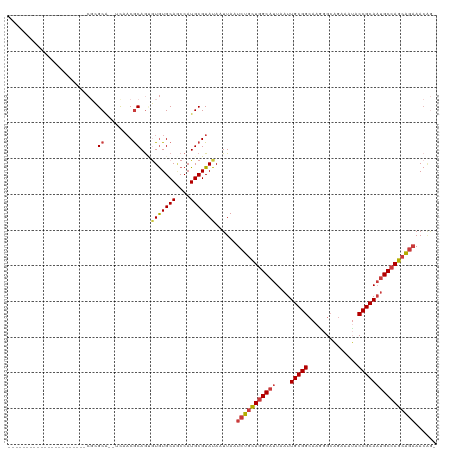
>dm3.chr2R 1720726 95 + 21146708 -CUGUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAUACGCAUGGCUGCACACACUGCGUUGA--UGCGCG---------------------- -.....((((((((((((((((((..........)))))))....))))))))))).........(((((.(.((((.....)))).).)--))))..---------------------- ( -39.20, z-score = -3.77, R) >droSim1.chr2R_random 462661 97 + 2996586 -CUGUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUCCCGCAAUUAAUGCCAGCAGAUUAAUUAUACGCAUGGCUGCACACGCUGCGUGGAGCUGCGCG---------------------- -.....((((((((((((((((..............)))))....))))))))))).........(((..((((.(((.......))).))))..)))---------------------- ( -39.14, z-score = -2.79, R) >droSec1.super_26 377489 97 + 826190 -CUGUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAUACGCAUGGCUGCACACGCUGCGUGAAGCUGCGCG---------------------- -.....((((((((((((((((((..........)))))))....))))))))))).........(((..((((.(((.......))).))))..)))---------------------- ( -41.00, z-score = -3.23, R) >droYak2.chr2L 1508450 95 - 22324452 -CUGUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAUACGCAUGGAUGCACACACUGCGUUGA--UGCGCG---------------------- -.....((((((((((((((((((..........)))))))....))))))))))).........(((((.((((((.....)))))).)--))))..---------------------- ( -43.60, z-score = -5.57, R) >droEre2.scaffold_4929 6019631 94 - 26641161 -CCGUUUCUGCUGGCAUUGCGGUAUUUCUGCCUUUGCCGCAAUUAAUGCCAGUAGAUUAAUUAUACGCAUGGUUGCACACACUGUGUUGG--UGC-CG---------------------- -.....((((((((((((((((((..........)))))))....)))))))))))..........((((.(..((((.....))))).)--)))-..---------------------- ( -34.70, z-score = -3.07, R) >droAna3.scaffold_13266 822418 95 - 19884421 -UUUUUACUGCUGGCAAUGCGGUUUUUCACCCAUUGCCGCAAUUAAUGCCAACAGAUUAAUUAUACGCAUGGUUGCACAUUCUGCGAUGGCGCGCU------------------------ -......(((.(((((.((((((............)))))).....))))).)))..........(((.(.((((((.....)))))).).)))..------------------------ ( -28.20, z-score = -1.32, R) >droWil1.scaffold_180700 4468269 120 - 6630534 CUGGUUUACGCUGGCGUUGCGGUUUUGCAGCCUUUGCCGCAAUUAAUGCUAACAGAUUAAUUAUACGCAUGGCUGCAUAAUUUGCGUUGGCUGGCUGGCUGGCUGUGUGUCUGUCUCCCU .........((..((((...((((.(((((((..(((...(((((((........)))))))....))).))))))).)))).))))..)).((..(((.(((.....))).)))..)). ( -39.70, z-score = -0.52, R) >consensus _CUGUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAUACGCAUGGCUGCACACACUGCGUUGA__UGCGCG______________________ ......((((((((((((((((((..........)))))))....)))))))))))........(((((((......))...)))))................................. (-27.35 = -28.17 + 0.82)

| Location | 1,720,726 – 1,720,821 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.85 |
| Shannon entropy | 0.35546 |
| G+C content | 0.49903 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
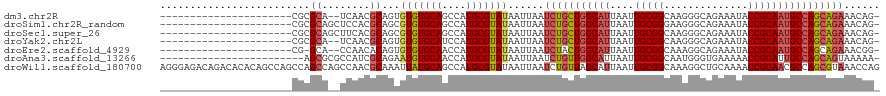
>dm3.chr2R 1720726 95 - 21146708 ----------------------CGCGCA--UCAACGCAGUGUGUGCAGCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAACAG- ----------------------.(((((--(....(((.....)))....))))))........(((((((((((....(((((..............)))))))))))))))).....- ( -34.64, z-score = -1.98, R) >droSim1.chr2R_random 462661 97 - 2996586 ----------------------CGCGCAGCUCCACGCAGCGUGUGCAGCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGGAAGGGCAGAAAUACCGCAAUGCCAGCAGAAACAG- ----------------------.((((((((.(((((...))))).)))..)))))........(((((((((((....(((((..............)))))))))))))))).....- ( -41.34, z-score = -3.51, R) >droSec1.super_26 377489 97 - 826190 ----------------------CGCGCAGCUUCACGCAGCGUGUGCAGCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAACAG- ----------------------.((((((((.(((((...))))).)))..)))))........(((((((((((....(((((..............)))))))))))))))).....- ( -40.04, z-score = -3.03, R) >droYak2.chr2L 1508450 95 + 22324452 ----------------------CGCGCA--UCAACGCAGUGUGUGCAUCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAACAG- ----------------------.(((((--(....(((.....)))....))))))........(((((((((((....(((((..............)))))))))))))))).....- ( -34.64, z-score = -2.50, R) >droEre2.scaffold_4929 6019631 94 + 26641161 ----------------------CG-GCA--CCAACACAGUGUGUGCAACCAUGCGUAUAAUUAAUCUACUGGCAUUAAUUGCGGCAAAGGCAGAAAUACCGCAAUGCCAGCAGAAACGG- ----------------------((-(((--(((......)).))))..................(((.(((((((....(((((..............)))))))))))).)))..)).- ( -25.74, z-score = -0.83, R) >droAna3.scaffold_13266 822418 95 + 19884421 ------------------------AGCGCGCCAUCGCAGAAUGUGCAACCAUGCGUAUAAUUAAUCUGUUGGCAUUAAUUGCGGCAAUGGGUGAAAAACCGCAUUGCCAGCAGUAAAAA- ------------------------.....((((..((((((((((((....)))))))......)))))))))....(((((((((((((((.....))).))))))).))))).....- ( -33.40, z-score = -2.26, R) >droWil1.scaffold_180700 4468269 120 + 6630534 AGGGAGACAGACACACAGCCAGCCAGCCAGCCAACGCAAAUUAUGCAGCCAUGCGUAUAAUUAAUCUGUUAGCAUUAAUUGCGGCAAAGGCUGCAAAACCGCAACGCCAGCGUAAACCAG .((..((((((......((..((......))....)).((((((((.((...))))))))))..)))))).((.....(((((((....)))))))....)).(((....)))...)).. ( -27.90, z-score = 0.38, R) >consensus ______________________CGCGCA__UCAACGCAGUGUGUGCAGCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAACAG_ .........................((........))...(((((((....)))))))......(((((((((((....(((((..............))))))))))))))))...... (-25.20 = -25.80 + 0.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:29 2011