
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,715,883 – 1,715,942 |
| Length | 59 |
| Max. P | 0.978623 |

| Location | 1,715,883 – 1,715,942 |
|---|---|
| Length | 59 |
| Sequences | 5 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 69.02 |
| Shannon entropy | 0.54619 |
| G+C content | 0.56789 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -13.71 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
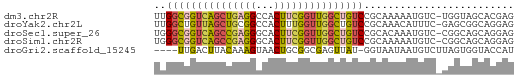
>dm3.chr2R 1715883 59 - 21146708 UUGGCGGUCAGCUGAGGCCACUUCGGUUGGCUGUCCGCAAAAAUGUC-UGGUAGCACGAG ..(((((((((((((((...))))))))))))))).((.........-.....))..... ( -20.44, z-score = -1.10, R) >droYak2.chr2L 1503802 59 + 22324452 UUGGCUGUUAGCUGCGGCCACUUUGGUUGGCUGUCCGCAAACAUUUC-GAGCGGCAGGAG .(((((((.....)))))))..........(((.((((.........-..)))))))... ( -19.50, z-score = -0.29, R) >droSec1.super_26 372755 59 - 826190 UGGGCGGUCAGCCGAGGGCACUUCGGUUGGCUGUCCGCACAAAUGUC-CGGCAGCAGGAG .((((((((((((((((...)))))))))))))))).........((-(.......))). ( -29.30, z-score = -3.14, R) >droSim1.chr2R 629957 59 - 19596830 UGGGCGGUCAGCCGAGGGCACUUCGGUUGGCUGUCCGCAAAAAUGUC-CGGCAGCAGGAG .((((((((((((((((...)))))))))))))))).........((-(.......))). ( -29.30, z-score = -3.41, R) >droGri2.scaffold_15245 8755951 55 - 18325388 ----UUGACUUACAAAGUAACUGCGGCGAGUUAU-GGUAAUAAUGUCUUAGUGGUACCAU ----.(((((..(...((....)).)..)))))(-((((...((......))..))))). ( -9.30, z-score = 0.17, R) >consensus UGGGCGGUCAGCCGAGGCCACUUCGGUUGGCUGUCCGCAAAAAUGUC_CGGCAGCAGGAG ..(((((((((((((((...)))))))))))))))......................... (-13.71 = -14.32 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:26 2011