| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,630,047 – 1,630,145 |
| Length | 98 |
| Max. P | 0.932648 |

| Location | 1,630,047 – 1,630,145 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 70.59 |
| Shannon entropy | 0.62016 |
| G+C content | 0.55044 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -11.45 |
| Energy contribution | -11.29 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
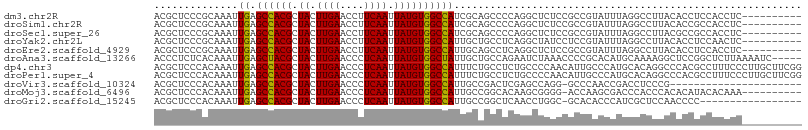
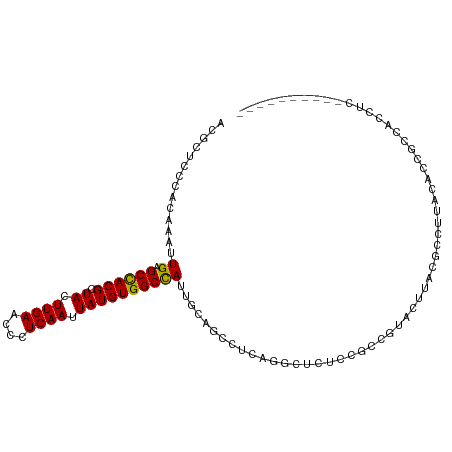
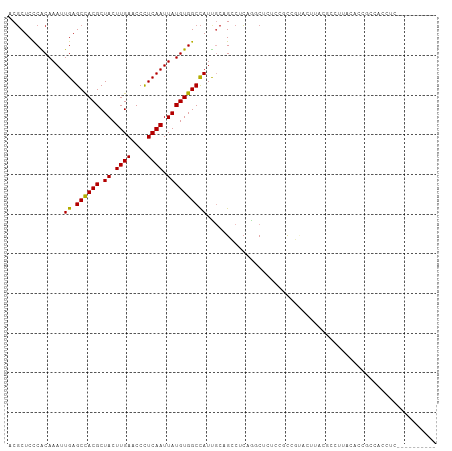
>dm3.chr2R 1630047 98 + 21146708 ACGCUCCCGCAAAUUGAGCCACGCUACUUGAACCUUCAAUUAUGUGGCCAUCGCAGCCCCAGGCUCUCCGCCGUAUUUAGGCCUUACACCUCCACCUC---------- ........((....((.((((((.((.((((....)))).))))))))))..))((((...))))....(((.......)))................---------- ( -20.80, z-score = -0.94, R) >droSim1.chr2R 531035 98 + 19596830 ACGCUCCCGCAAAUUGAGCCACGCUACUUGAACCUUCAAUUAUGUGGCCAUCGCAGCCCCAGGCUCUCCGCCGUAUUUAGGCCUUACACCGCCACCUC---------- ..(((...((....((.((((((.((.((((....)))).))))))))))..)))))....(((.....))).......(((........))).....---------- ( -22.90, z-score = -1.19, R) >droSec1.super_26 266714 98 + 826190 ACGCUCCCGCAAAUUGAGCCACGCUACUUGAACCUUCAAUUAUGUGGCCAUCGCAGCCCCAGGCUCUCCGCCGUAUUUAGGCCUUACGCCGCCACCUC---------- ..((....((....((.((((((.((.((((....)))).))))))))))..)).......(((.....(((.......))).....)))))......---------- ( -24.20, z-score = -1.04, R) >droYak2.chr2L 1400548 98 - 22324452 ACGCUCCCGCAAAUUGAGCCACGCUACUUGAACCUUCAAUUAUGUGGCCAUUGCUGCCUCAGGCUAUCCUCCGUAUUUAGGCCUUACACCUCCAACUC---------- ........((((..((.((((((.((.((((....)))).)))))))))))))).((((...((........))....))))................---------- ( -20.30, z-score = -1.24, R) >droEre2.scaffold_4929 5907493 98 - 26641161 ACGCUCCCGCAAAUUGAGCCACGCUACUUGAACCUUCAAUUAUGUGGCCAUUGCAGCCUCAGGCUCUCCGCCGUAUUUAGGCCUUACACCUCCACCUC---------- ........((((..((.((((((.((.((((....)))).)))))))))))))).((((..(((.....)))......))))................---------- ( -24.30, z-score = -2.11, R) >droAna3.scaffold_13266 680774 103 - 19884421 ACCCUCUCACAAAUUGAGCUACGCUACUUGAACCCUCAAUUAUGUGGCUAUUGCUGCCAGAAUCUAAACCCCGCACAUGCAAAAGGCUCCGGCUCUUAAAAUC----- ................(((((((.((.((((....)))).)))))))))......(((.((..((.......((....))...))..)).)))..........----- ( -17.50, z-score = -0.53, R) >dp4.chr3 4681374 108 - 19779522 ACGCUCCCACAAAUUGAGCCACGCUACUUGAACCCUCAAUUAUGUGGCCAUUUCUGCCUCUGCCCCAACAUUGCCCAUGCACAGGCCCACGCCUUUCCCUUGCUUCGG ..((..........((.((((((.((.((((....)))).)))))))))).....((((.(((...............))).))))...............))..... ( -20.56, z-score = -0.70, R) >droPer1.super_4 3952786 108 + 7162766 ACGCUCCCACAAAUUGAGCCACGCUACUUGAACCCUCAAUUAUGUGGCCAUUUCUGCCUCUGCCCCAACAUUGCCCAUGCACAGGCCCACGCCUUUCCCUUGCUUCGG ..((..........((.((((((.((.((((....)))).)))))))))).....((((.(((...............))).))))...............))..... ( -20.56, z-score = -0.70, R) >droVir3.scaffold_10324 768737 85 + 1288806 ACGCUCCCACAAAUUGAGCCACGCUACUUGAACCCUCAAUUAUGUGGCCAUUGCCGACUCGAGCCAGG-GCCCAACCGACCUCCCG---------------------- ..((((........((.((((((.((.((((....)))).))))))))))..........))))..((-(............))).---------------------- ( -18.67, z-score = -1.33, R) >droMoj3.scaffold_6496 2612153 97 + 26866924 ACGCUCCCACAAAUUGAGCCACGCUACUUGAACCCUCAAUUAUGUGGCCAUUGCCGGCACAAGCGGGG-ACCAAGCGACCCACCCACACAUACACAAA---------- .(((((((.....(((.((((((.((.((((....)))).)))))(((....)))))).)))...)))-)....))).....................---------- ( -22.40, z-score = -1.07, R) >droGri2.scaffold_15245 8466661 90 + 18325388 ACGCUCCCACAAAUUGAGCCACGCUACUUGAACCCUCAAUUAUGUGGCCAUUGCCGGCUCAACCUGGC-GCACACCCAUCGCUCCAACCCC----------------- .............((((((((((.((.((((....)))).)))))(((....))))))))))..(((.-((.........)).))).....----------------- ( -21.40, z-score = -2.24, R) >consensus ACGCUCCCACAAAUUGAGCCACGCUACUUGAACCCUCAAUUAUGUGGCCAUUGCAGCCUCAGGCUCUCCGCCGUACUUACGCCUUACACCGCCACCUC__________ ..............((.((((((.((.((((....)))).)))))))))).......................................................... (-11.45 = -11.29 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:20 2011