| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,167,730 – 2,167,826 |
| Length | 96 |
| Max. P | 0.925048 |

| Location | 2,167,730 – 2,167,826 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 66.68 |
| Shannon entropy | 0.64011 |
| G+C content | 0.46807 |
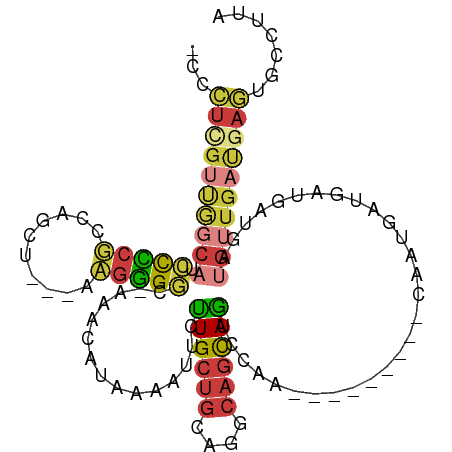
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -14.97 |
| Energy contribution | -15.67 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
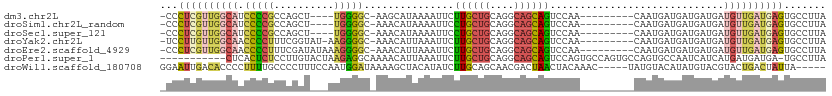
>dm3.chr2L 2167730 96 - 23011544 -CCCUCGUUGGCAUCCCCGCCAGCU----UGGGGC-AAGCAUAAAAUUCUUGCUGCAGGCAGCAGUCCAA---------CAAUGAUGAUGAUGAUGUUGAUGAGUGCCUUA -..(((((..(((((...((..(((----((.(((-(((.(......))))))).))))).)).(((...---------....)))......)))))..)))))....... ( -32.00, z-score = -1.04, R) >droSim1.chr2L_random 114598 96 - 909653 -CCCUCGUUGGCAUCCCCGCCAGCU----UGGGGC-AAACAUAAAAUUCCUGCUGCAGGCAGCAGUCCAA---------CAAUGAUGAUGAUGAUGUUGAUGAGUGCCUUA -.((..((((((......)))))).----.))(((-(..(((.......((((((....))))))..(((---------((.(.((....)).)))))))))..))))... ( -32.80, z-score = -1.72, R) >droSec1.super_121 35854 96 - 59987 -CCCUCGUUGGCAUCCCCGCCAGCU----UGGGGC-AAACAUAAAAUUCCUGCUGCAGGCAGCAGUCCAA---------CAAUGAUGAUGAUGAUGUUGAUGAGUGCCUUA -.((..((((((......)))))).----.))(((-(..(((.......((((((....))))))..(((---------((.(.((....)).)))))))))..))))... ( -32.80, z-score = -1.72, R) >droYak2.chr2L 2139549 99 - 22324452 -UCCUUGUUGGCAACCCCUUUCGGUAU-AAGGGGC-AAACAUUAAAUUCUUGCUGCAGGCAGCAGUCCAA---------CAAUGAUGAUGAUGAUGUUGAUGAGUGCCUUA -..((..(..(((.((((((.......-)))))).-...(((((......(((((....)))))(((...---------....)))..))))).)))..)..))....... ( -27.00, z-score = -0.47, R) >droEre2.scaffold_4929 2202445 100 - 26641161 -CCCUCGUUGGCAACCCCUUUCGAUAUAAAGGGGC-AAACAUUAAAUUCUUGCUGCAGGCAGCAGUCCAA---------CAAUGAUGAUGAUGAUGUUGAUGAGUGCCUUA -..(((((..(((.(((((((......))))))).-...(((((......(((((....)))))(((...---------....)))..))))).)))..)))))....... ( -30.30, z-score = -1.89, R) >droPer1.super_1 4529299 99 + 10282868 -----------CUCACUCUCCUUGUACUAAGAGGCAAAACAUUAAAUUCUUGCUGCAGGCAGCAGUCCAGUGCCAGUGCCAGUGCCAAUCAUCAUGAUGAUGA-UGCCUUA -----------............(((((....((((...((((......((((((....))))))...))))....)))))))))..((((((.....)))))-)...... ( -22.70, z-score = 0.50, R) >droWil1.scaffold_180708 10439305 101 + 12563649 GGAAUUGACACCCCUUUUGCCCCUUUCCAAUGGAUAAAAGCUACAUAUCUUGCAGCAACGACUAACUACAAAC-----UAUGUACAUAUGUACGUACUGACUAUUA----- ((((....................))))(((((......(((.((.....)).))).................-----(((((((....)))))))....))))).----- ( -14.75, z-score = -1.12, R) >consensus _CCCUCGUUGGCAUCCCCGCCAGCU___AAGGGGC_AAACAUAAAAUUCUUGCUGCAGGCAGCAGUCCAA_________CAAUGAUGAUGAUGAUGUUGAUGAGUGCCUUA ...((((((((((.(((((..........)))))...............((((((....)))))).............................))))))))))....... (-14.97 = -15.67 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:24 2011