| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,627,669 – 1,627,719 |
| Length | 50 |
| Max. P | 0.751481 |

| Location | 1,627,669 – 1,627,719 |
|---|---|
| Length | 50 |
| Sequences | 4 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 50.00 |
| Shannon entropy | 0.80999 |
| G+C content | 0.40015 |
| Mean single sequence MFE | -14.52 |
| Consensus MFE | -2.50 |
| Energy contribution | -1.44 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.86 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.17 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.751481 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 1627669 50 - 21146708 UUCAACGGAUCUAGUAUACCUUUUUA-------CUCAAAGAGUAACGGGUAUAACAA ..............(((((((..(((-------(((...)))))).))))))).... ( -11.30, z-score = -2.65, R) >droSim1.chr2R 528067 55 - 19596830 UUCAACUGAUGCAGGUUGGUCCCUUCGUAGA-CCAAAUCGCAGAAUAAGUGGCAAA- ....(((..(((...((((((........))-))))...))).....)))......- ( -12.10, z-score = -0.71, R) >droSec1.super_26 263671 55 - 826190 UUCAACUGAUGCAGGUUGGUCCCCUUGUGGA-CCAAAUCGCAGAAAAAGUGGCAAA- ....(((..(((...(((((((......)))-))))...))).....)))......- ( -15.40, z-score = -1.23, R) >droGri2.scaffold_14906 3787607 57 - 14172833 UCGAACAUAACCAUUAUACCCUCUUAACCCAUUUUGAAUGGGUUUUGGGUAUAAAAA .............((((((((....(((((((.....)))))))..))))))))... ( -19.30, z-score = -4.35, R) >consensus UUCAACUGAUCCAGGAUACCCCCUUAGU_GA_CCAAAACGCAGAAUAAGUAGAAAA_ ..............(((((((.........................))))))).... ( -2.50 = -1.44 + -1.06)

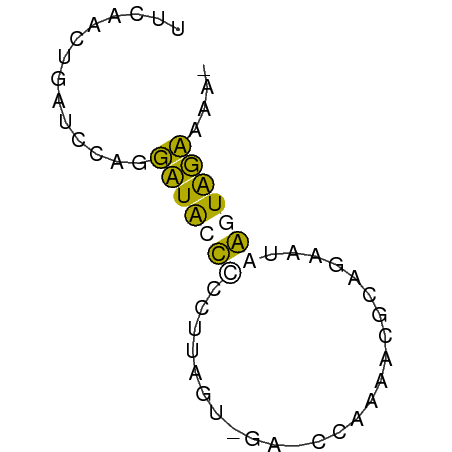
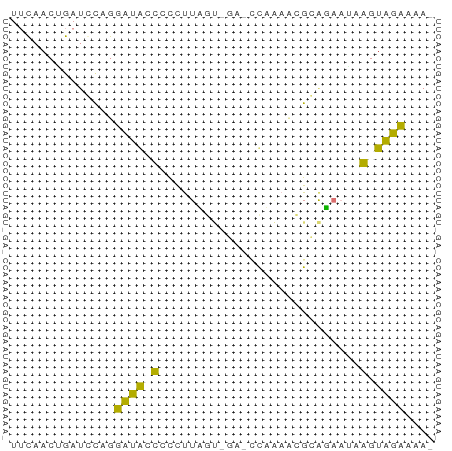
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:19 2011