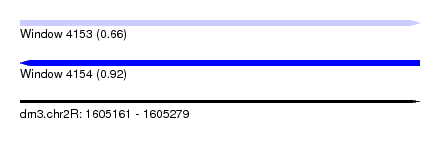
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,605,161 – 1,605,279 |
| Length | 118 |
| Max. P | 0.923300 |

| Location | 1,605,161 – 1,605,279 |
|---|---|
| Length | 118 |
| Sequences | 13 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.51 |
| Shannon entropy | 0.30769 |
| G+C content | 0.52586 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -27.54 |
| Energy contribution | -28.87 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1605161 118 + 21146708 UUGUUAUUUGCACCUGCAGCACUGCCACAUUGAUUGCUCCCUUGAUGUGCUACAGGUGGUGUUGGACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUAUAUUU ...............(((((((((.(((((..(........)..)))))...)))..(((((.(((......)))...)))))((((.((((....)))).))))))))))....... ( -36.60, z-score = -1.80, R) >droSim1.chr2R 502535 118 + 19596830 UUGUUAUUUGCACCUGCAGCACUGCCACAUUGAUUGCUCCCUUGAUGUGCUACAGGUGGUGUUGGACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUAUAUUU ...............(((((((((.(((((..(........)..)))))...)))..(((((.(((......)))...)))))((((.((((....)))).))))))))))....... ( -36.60, z-score = -1.80, R) >droSec1.super_26 242230 118 + 826190 UUGUUAUUUGCACCUGCAGCACUGCCACAUUGAUUGCUCCCUUGAUGUGCUACAGGUGGUGUUGGACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUAUAUUU ...............(((((((((.(((((..(........)..)))))...)))..(((((.(((......)))...)))))((((.((((....)))).))))))))))....... ( -36.60, z-score = -1.80, R) >droYak2.chr2L 1378843 118 - 22324452 UUGUUAUUUGCACCUGCAGCACUGCCACAUUGAUUGCUCCCUUGAUGUGCUACAGGUGGUGUUGGACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUAUAUUU ...............(((((((((.(((((..(........)..)))))...)))..(((((.(((......)))...)))))((((.((((....)))).))))))))))....... ( -36.60, z-score = -1.80, R) >droEre2.scaffold_4929 5885046 118 - 26641161 UUGUUAUUUGCACCUGCAGCACUGCCACAUUGAUUGCUCCCUUGAUGUGCUACAGGUGGUGUUGGACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUAUAUUU ...............(((((((((.(((((..(........)..)))))...)))..(((((.(((......)))...)))))((((.((((....)))).))))))))))....... ( -36.60, z-score = -1.80, R) >droAna3.scaffold_13266 660115 118 - 19884421 UUGGUACUUGCACCUGCAGCACUGCCACAGUGGUUGCUCCCUUGAUGUGCUACAGGUGGUGUUGGACUUUUUUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUGCAAUA ...(((...(((((((.(((((...((.((.((.....))))))..))))).)))))(((((.(((......)))...)))))((((.((((....)))).)))).))...))).... ( -39.40, z-score = -0.30, R) >dp4.chr3 4664118 118 - 19779522 UUGUUAUUUGCACCUGCAGCACUGCCACAUUGAUUGCUCCCCUGAUGUGCUACAGGUGGUGUUGGACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUGUAAUU .......(((((...(((((((((.((((((............))))))...)))..(((((.(((......)))...)))))((((.((((....)))).))))))))))))))).. ( -36.20, z-score = -1.30, R) >droPer1.super_4 3935692 118 + 7162766 UUGUUAUUUGCACCUGCAGCACUGCCACAUUGAUUGCUCCCCUGAUGUGCUACAGGUGGUGUUGGACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUGUAAUU .......(((((...(((((((((.((((((............))))))...)))..(((((.(((......)))...)))))((((.((((....)))).))))))))))))))).. ( -36.20, z-score = -1.30, R) >droWil1.scaffold_180569 1262531 118 + 1405142 UUGUUAUUUGCACCUGCAGCACUGCCACAUUGAUUGCUCCCUUGAUGUGCUACAGGUGGUGUUGGACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUGUAAUU .......(((((...(((((((((.(((((..(........)..)))))...)))..(((((.(((......)))...)))))((((.((((....)))).))))))))))))))).. ( -38.50, z-score = -2.07, R) >droVir3.scaffold_10324 746095 118 + 1288806 UUGUUAUUUGCACCUGCAGCACUGCCGCAUUGAUUGCUCCCUUGAUGUGCUACAGGUGGUGUUGGACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUGCAGGG ............((((((((((((.(((((..(........)..)))))...)))..(((((.(((......)))...)))))((((.((((....)))).))))...))))))))). ( -44.30, z-score = -2.01, R) >droMoj3.scaffold_6496 2586451 118 + 26866924 UUAUUAUUUGCACCUGCAGCACUGCCGCACUGAUUGCUCCCUUGAUGUGCUACAGGUGGUGUUAAACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUGUAAAG ......((((((...(((((((((..((((................))))..)))..(((((................)))))((((.((((....)))).)))))))))))))))). ( -34.78, z-score = -1.50, R) >droGri2.scaffold_15245 8447784 118 + 18325388 UUGUUAUUUGCACCUGCAGCACUGCCGCAUUGAUUGCUCCCUUGAUGUGCUACAGGUGGUGUUGGACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUGCAAAU .....(((((((...(((((((((.(((((..(........)..)))))...)))..(((((.(((......)))...)))))((((.((((....)))).))))))))))))))))) ( -41.50, z-score = -2.30, R) >anoGam1.chr3R 22936447 93 + 53272125 -------------------GGUUGUUGUUGUUGCUGUUGAGCUGGCAGGCGUUCGACAGGUCCGGGCUGCGCUCCC--GCGGCGG-CCCCAGAAACCGAGCACUCGGGUGCUGUG--- -------------------((((.(((......(((((((((........)))))))))....(((((((((....--)).))))-)))))).)))).(((((....)))))...--- ( -39.70, z-score = -0.10, R) >consensus UUGUUAUUUGCACCUGCAGCACUGCCACAUUGAUUGCUCCCUUGAUGUGCUACAGGUGGUGUUGGACUUUUCUCCCUUGCGCCGGGCCCAAGAAAUCUUGCGCUCUGCUGCUGUAAUU .........(((((((.(((((........................))))).)))))(((((.(((......)))...)))))((((.((((....)))).)))).)).......... (-27.54 = -28.87 + 1.33)



| Location | 1,605,161 – 1,605,279 |
|---|---|
| Length | 118 |
| Sequences | 13 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.51 |
| Shannon entropy | 0.30769 |
| G+C content | 0.52586 |
| Mean single sequence MFE | -39.51 |
| Consensus MFE | -32.11 |
| Energy contribution | -33.07 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1605161 118 - 21146708 AAAUAUAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUCCAACACCACCUGUAGCACAUCAAGGGAGCAAUCAAUGUGGCAGUGCUGCAGGUGCAAAUAACAA .......((.((.(.((.((((....)))).)).).))))...(((......))).....((((((((((((.(((...((....))....)))..)))))))))))).......... ( -40.20, z-score = -2.40, R) >droSim1.chr2R 502535 118 - 19596830 AAAUAUAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUCCAACACCACCUGUAGCACAUCAAGGGAGCAAUCAAUGUGGCAGUGCUGCAGGUGCAAAUAACAA .......((.((.(.((.((((....)))).)).).))))...(((......))).....((((((((((((.(((...((....))....)))..)))))))))))).......... ( -40.20, z-score = -2.40, R) >droSec1.super_26 242230 118 - 826190 AAAUAUAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUCCAACACCACCUGUAGCACAUCAAGGGAGCAAUCAAUGUGGCAGUGCUGCAGGUGCAAAUAACAA .......((.((.(.((.((((....)))).)).).))))...(((......))).....((((((((((((.(((...((....))....)))..)))))))))))).......... ( -40.20, z-score = -2.40, R) >droYak2.chr2L 1378843 118 + 22324452 AAAUAUAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUCCAACACCACCUGUAGCACAUCAAGGGAGCAAUCAAUGUGGCAGUGCUGCAGGUGCAAAUAACAA .......((.((.(.((.((((....)))).)).).))))...(((......))).....((((((((((((.(((...((....))....)))..)))))))))))).......... ( -40.20, z-score = -2.40, R) >droEre2.scaffold_4929 5885046 118 + 26641161 AAAUAUAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUCCAACACCACCUGUAGCACAUCAAGGGAGCAAUCAAUGUGGCAGUGCUGCAGGUGCAAAUAACAA .......((.((.(.((.((((....)))).)).).))))...(((......))).....((((((((((((.(((...((....))....)))..)))))))))))).......... ( -40.20, z-score = -2.40, R) >droAna3.scaffold_13266 660115 118 + 19884421 UAUUGCAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAAAAAAGUCCAACACCACCUGUAGCACAUCAAGGGAGCAACCACUGUGGCAGUGCUGCAGGUGCAAGUACCAA ..((((....)))).((.((((....)))).))..((..(...(((......))).....((((((((((((.(((((((.....)).)).)))..))))))))))))...)..)).. ( -41.10, z-score = -1.03, R) >dp4.chr3 4664118 118 + 19779522 AAUUACAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUCCAACACCACCUGUAGCACAUCAGGGGAGCAAUCAAUGUGGCAGUGCUGCAGGUGCAAAUAACAA .......((.((.(.((.((((....)))).)).).))))...(((......))).....((((((((((((.(((.(.((....))..).)))..)))))))))))).......... ( -39.30, z-score = -1.65, R) >droPer1.super_4 3935692 118 - 7162766 AAUUACAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUCCAACACCACCUGUAGCACAUCAGGGGAGCAAUCAAUGUGGCAGUGCUGCAGGUGCAAAUAACAA .......((.((.(.((.((((....)))).)).).))))...(((......))).....((((((((((((.(((.(.((....))..).)))..)))))))))))).......... ( -39.30, z-score = -1.65, R) >droWil1.scaffold_180569 1262531 118 - 1405142 AAUUACAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUCCAACACCACCUGUAGCACAUCAAGGGAGCAAUCAAUGUGGCAGUGCUGCAGGUGCAAAUAACAA .......((.((.(.((.((((....)))).)).).))))...(((......))).....((((((((((((.(((...((....))....)))..)))))))))))).......... ( -40.20, z-score = -2.19, R) >droVir3.scaffold_10324 746095 118 - 1288806 CCCUGCAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUCCAACACCACCUGUAGCACAUCAAGGGAGCAAUCAAUGCGGCAGUGCUGCAGGUGCAAAUAACAA ((((...((.((.(.((.((((....)))).)).).)))).))))...............((((((((((((.((....))(((.....)))....)))))))))))).......... ( -42.90, z-score = -1.73, R) >droMoj3.scaffold_6496 2586451 118 - 26866924 CUUUACAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUUUAACACCACCUGUAGCACAUCAAGGGAGCAAUCAGUGCGGCAGUGCUGCAGGUGCAAAUAAUAA ((((...((.((.(.((.((((....)))).)).).))))...)))).............((((((((((((.((....))(((.....)))....)))))))))))).......... ( -38.80, z-score = -1.68, R) >droGri2.scaffold_15245 8447784 118 - 18325388 AUUUGCAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUCCAACACCACCUGUAGCACAUCAAGGGAGCAAUCAAUGCGGCAGUGCUGCAGGUGCAAAUAACAA ((((((.((.((.(.((.((((....)))).)).).))))...(((......)))......(((((((((((.((....))(((.....)))....)))))))))))))))))..... ( -42.50, z-score = -2.16, R) >anoGam1.chr3R 22936447 93 - 53272125 ---CACAGCACCCGAGUGCUCGGUUUCUGGGG-CCGCCGC--GGGAGCGCAGCCCGGACCUGUCGAACGCCUGCCAGCUCAACAGCAACAACAACAACC------------------- ---..((((((....))))).)(((.((((((-(.((.((--....)))).)))).((.(((.((......)).))).))..))).)))..........------------------- ( -28.50, z-score = 1.06, R) >consensus AAUUACAGCAGCAGAGCGCAAGAUUUCUUGGGCCCGGCGCAAGGGAGAAAAGUCCAACACCACCUGUAGCACAUCAAGGGAGCAAUCAAUGUGGCAGUGCUGCAGGUGCAAAUAACAA .......((.((.(.((.((((....)))).)).).))))...(((......))).....((((((((((((.......((....)).........)))))))))))).......... (-32.11 = -33.07 + 0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:16 2011