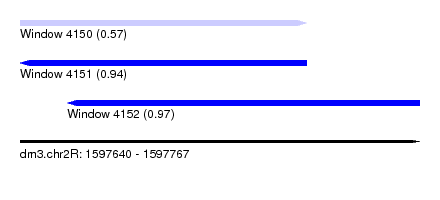
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,597,640 – 1,597,767 |
| Length | 127 |
| Max. P | 0.971557 |

| Location | 1,597,640 – 1,597,731 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 89.93 |
| Shannon entropy | 0.19229 |
| G+C content | 0.38237 |
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -13.91 |
| Energy contribution | -14.01 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
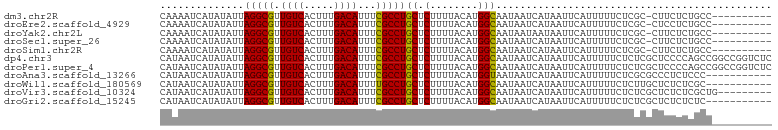
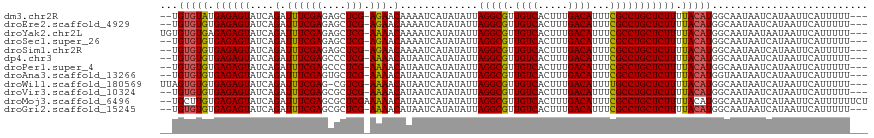
>dm3.chr2R 1597640 91 + 21146708 ----------GGCAGAGAAG-GCGAGAAAAAUGAAUUAUGAUUAUUGCCAUGUAAAAGAGCAGGCGAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUUUG ----------........((-(((....................(((((.(((......))))))))..((((.....)))).))))).............. ( -20.20, z-score = -2.45, R) >droEre2.scaffold_4929 5876597 91 - 26641161 ----------GGCAGAGGAG-GCGAGAAAAAUGAAUUAUGAUUAUUGCCAUGUAAAAGAGCAGGCGAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUUUG ----------..((.(..((-(((....................(((((.(((......))))))))..((((.....)))).)))))..)...))...... ( -20.70, z-score = -2.37, R) >droYak2.chr2L 1368484 91 - 22324452 ----------GGCAGAGAAG-GCGAGAAAAAUGAAUUAUUAUUAUUGCCAUGUAAAAGAGCAGGCGAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUUUG ----------........((-(((....................(((((.(((......))))))))..((((.....)))).))))).............. ( -20.20, z-score = -2.45, R) >droSec1.super_26 235123 91 + 826190 ----------GGCAGAGAAG-GCGAGAAAAAUGAAUUAUGAUUAUUGCCAUGUAAAAGAGCAGGCGAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUUUG ----------........((-(((....................(((((.(((......))))))))..((((.....)))).))))).............. ( -20.20, z-score = -2.45, R) >droSim1.chr2R 495083 91 + 19596830 ----------GGCAGAGAAG-GCGAGAAAAAUGAAUUAUGAUUAUUGCCAUGUAAAAGAGCAGGCGAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUUUG ----------........((-(((....................(((((.(((......))))))))..((((.....)))).))))).............. ( -20.20, z-score = -2.45, R) >dp4.chr3 4656331 102 - 19779522 GAGACCGGCCGGCUGGGGAGCGAGAGAAAAAUGAAUUAUGAUUAUUGCCAUGUAAAAGAGCAGGCGAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUAUG ....((((....))))((.(((......................(((((.(((......))))))))..((((.....)))).))))).............. ( -21.90, z-score = -1.13, R) >droPer1.super_4 3927886 102 + 7162766 GAGACCGGCCGGCUGGGGAGCGAGAGAAAAAUGAAUUAUGAUUAUUGCCAUGUAAAAGAGCAGGCGAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUAUG ....((((....))))((.(((......................(((((.(((......))))))))..((((.....)))).))))).............. ( -21.90, z-score = -1.13, R) >droAna3.scaffold_13266 652865 91 - 19884421 -----------GGGAGAGGGCGCGAGAAAAAUGAAUUAUGAUUAUUACCAUGUAAAAGAGCAGGCGAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUAUG -----------.........(....)..........(((((((((......((......))(((((...((((.....)))).))))).....))))))))) ( -14.70, z-score = -0.55, R) >droWil1.scaffold_180569 1251445 91 + 1405142 -----------GCGAGAGAGCAAGAGAAAAAUGAAUUAUGAUUAUUGCCAUGUAAAAGAGCAGGCAAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUAUG -----------((......))............((((((((((((((((.(((......))))))))..((((.....)))).....)))).)))))))... ( -17.10, z-score = -2.11, R) >droVir3.scaffold_10324 734206 93 + 1288806 ---------CAGCGAGAGAGCGAGAGAAAAAUGAAUUAUGAUUAUUGCCAUGUAAAAGAGCAGGCGAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUAUG ---------..((......))...............(((((((((.(((........).))(((((...((((.....)))).))))).....))))))))) ( -17.60, z-score = -2.08, R) >droGri2.scaffold_15245 8436806 91 + 18325388 -----------GAGAGAGAGCGAGAGAAAAAUGAAUUAUGAUUAUUGCCAUGUAAAAGAGCAGGCGAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUAUG -----------.....((.(((......................(((((.(((......))))))))..((((.....)))).))))).............. ( -16.50, z-score = -2.45, R) >consensus __________GGCAGGGGAGCGCGAGAAAAAUGAAUUAUGAUUAUUGCCAUGUAAAAGAGCAGGCGAAAUGUCAAAGUGACAACGCCUAAUAUAUGAUUAUG .................................((((((((((((((((.(((......))))))))..((((.....)))).....)))).)))))))... (-13.91 = -14.01 + 0.10)

| Location | 1,597,640 – 1,597,731 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 89.93 |
| Shannon entropy | 0.19229 |
| G+C content | 0.38237 |
| Mean single sequence MFE | -15.06 |
| Consensus MFE | -13.31 |
| Energy contribution | -13.15 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1597640 91 - 21146708 CAAAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUUCUCGC-CUUCUCUGCC---------- .............((((((.((((.....))))...))))))...........(((.......................))-).........---------- ( -15.30, z-score = -2.32, R) >droEre2.scaffold_4929 5876597 91 + 26641161 CAAAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUUCUCGC-CUCCUCUGCC---------- .............((((((.((((.....))))...))))))...........(((.......................))-).........---------- ( -15.30, z-score = -2.28, R) >droYak2.chr2L 1368484 91 + 22324452 CAAAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUAAUAAUUCAUUUUUCUCGC-CUUCUCUGCC---------- .............((((((.((((.....))))...))))))...........(((.......................))-).........---------- ( -15.30, z-score = -2.25, R) >droSec1.super_26 235123 91 - 826190 CAAAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUUCUCGC-CUUCUCUGCC---------- .............((((((.((((.....))))...))))))...........(((.......................))-).........---------- ( -15.30, z-score = -2.32, R) >droSim1.chr2R 495083 91 - 19596830 CAAAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUUCUCGC-CUUCUCUGCC---------- .............((((((.((((.....))))...))))))...........(((.......................))-).........---------- ( -15.30, z-score = -2.32, R) >dp4.chr3 4656331 102 + 19779522 CAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUUCUCUCGCUCCCCAGCCGGCCGGUCUC .............((((((.((((.....))))...))))))..........((((..((....))...............(((....)))..))))..... ( -16.80, z-score = -0.82, R) >droPer1.super_4 3927886 102 - 7162766 CAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUUCUCUCGCUCCCCAGCCGGCCGGUCUC .............((((((.((((.....))))...))))))..........((((..((....))...............(((....)))..))))..... ( -16.80, z-score = -0.82, R) >droAna3.scaffold_13266 652865 91 + 19884421 CAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGUAAUAAUCAUAAUUCAUUUUUCUCGCGCCCUCUCCC----------- .............((((((.((((.....))))...)))))).........(((((....)))))..........................----------- ( -14.80, z-score = -2.40, R) >droWil1.scaffold_180569 1251445 91 - 1405142 CAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUUGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUUCUCUUGCUCUCUCGC----------- .............((((((.((((.....))))...))))))...........(((((.....................))))).......----------- ( -12.90, z-score = -1.23, R) >droVir3.scaffold_10324 734206 93 - 1288806 CAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUUCUCUCGCUCUCUCGCUG--------- ..............(((((.((((.....))))...)))))((.(........))).........................((......))..--------- ( -14.40, z-score = -2.25, R) >droGri2.scaffold_15245 8436806 91 - 18325388 CAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUUCUCUCGCUCUCUCUC----------- ..............(((((.((((.....))))...)))))((.(........)))...................................----------- ( -13.50, z-score = -2.66, R) >consensus CAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUUCUCGCGCUCCCCUGCC__________ ..............(((((.((((.....))))...)))))((.(........))).............................................. (-13.31 = -13.15 + -0.16)
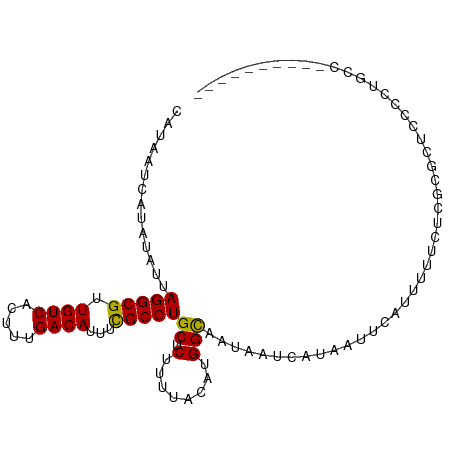
| Location | 1,597,655 – 1,597,767 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Shannon entropy | 0.09496 |
| G+C content | 0.35161 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.07 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1597655 112 - 21146708 --UGUGUAUGAGAGUAUCAGAUUUCGAGAGCUCG-AGAACAAAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUU--- --.(((((.((((((....(.((((((....)))-))).).............(((((.((((.....))))...))))))))))).))))).......................--- ( -29.90, z-score = -2.86, R) >droEre2.scaffold_4929 5876612 112 + 26641161 --UGUGUGUGAGAGUAUCAGAUUUCGAGAGCUCG-AGAACAAAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUU--- --.(((((.((((((....(.((((((....)))-))).).............(((((.((((.....))))...))))))))))).))))).......................--- ( -29.60, z-score = -2.37, R) >droYak2.chr2L 1368499 114 + 22324452 UGUGUGUGAGAGAGUAUCAGAUUUCGAGAGCUCG-AAAACAAAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUAAUAAUUCAUUUUU--- (((((((((((((((....(.((((((....)))-))).).............(((((.((((.....))))...))))))))))))))))).)))...................--- ( -33.50, z-score = -3.78, R) >droSec1.super_26 235138 112 - 826190 --UGUGUGUGAGAGUAUCAGAUUUCGAGAGCUCG-AGAACAAAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUU--- --.(((((.((((((....(.((((((....)))-))).).............(((((.((((.....))))...))))))))))).))))).......................--- ( -29.60, z-score = -2.37, R) >droSim1.chr2R 495098 112 - 19596830 --UGUGUGUGAGAGUAUCAGAUUUCGAGAGCUCG-AGAACAAAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUU--- --.(((((.((((((....(.((((((....)))-))).).............(((((.((((.....))))...))))))))))).))))).......................--- ( -29.60, z-score = -2.37, R) >dp4.chr3 4656357 112 + 19779522 --UGUGUGUGAGAGUAUCAGAUUUCGAGCCCUCG-AAAACAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUU--- --.(((((.((((((....(.((((((....)))-))).).............(((((.((((.....))))...))))))))))).))))).......................--- ( -29.50, z-score = -3.15, R) >droPer1.super_4 3927912 112 - 7162766 --UGUGUGUGAGAGUAUCAGAUUUCGAGCCCUCG-AAAACAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUU--- --.(((((.((((((....(.((((((....)))-))).).............(((((.((((.....))))...))))))))))).))))).......................--- ( -29.50, z-score = -3.15, R) >droAna3.scaffold_13266 652880 112 + 19884421 --UGUGUGUGAGAGUAUCAGAUUUCGAGUGCUCG-AAAACAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGUAAUAAUCAUAAUUCAUUUUU--- --.(((((.((((((....(.((((((....)))-))).).............(((((.((((.....))))...))))))))))).))))).......................--- ( -29.50, z-score = -2.86, R) >droWil1.scaffold_180569 1251460 113 - 1405142 UUAGUGUGUGAGAGUAUCAGAUUUCGAG-CGUCG-AAAACAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUUGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUU--- ...(((((.((((((....(.(((((..-...))-))).).............(((((.((((.....))))...))))))))))).))))).......................--- ( -26.40, z-score = -1.87, R) >droVir3.scaffold_10324 734223 112 - 1288806 --UGUGUGUGAGAGUAUCAGAUUUCGAGCGCUCG-AAAACAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUU--- --.(((((.((((((....(.((((((....)))-))).).............(((((.((((.....))))...))))))))))).))))).......................--- ( -29.50, z-score = -2.66, R) >droMoj3.scaffold_6496 2573716 116 - 26866924 --UGCUUGUGAGAGUAUCAGAUUUCGAGCGCUCGAAAAACAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUUUCU --((((((((((((.......((((((....))))))...............((((((.((((.....))))...))))))..)))))))).))))...................... ( -29.90, z-score = -3.04, R) >droGri2.scaffold_15245 8436821 112 - 18325388 --UGUGUGUGAGAGUAUCAGAUUUCGAGCGCUCG-AAAACAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUU--- --.(((((.((((((....(.((((((....)))-))).).............(((((.((((.....))))...))))))))))).))))).......................--- ( -29.50, z-score = -2.66, R) >consensus __UGUGUGUGAGAGUAUCAGAUUUCGAGAGCUCG_AAAACAUAAUCAUAUAUUAGGCGUUGUCACUUUGACAUUUCGCCUGCUCUUUUACAUGGCAAUAAUCAUAAUUCAUUUUU___ ...(((((.((((((....(.((((((....))).))).).............(((((.((((.....))))...))))))))))).))))).......................... (-25.27 = -25.07 + -0.21)
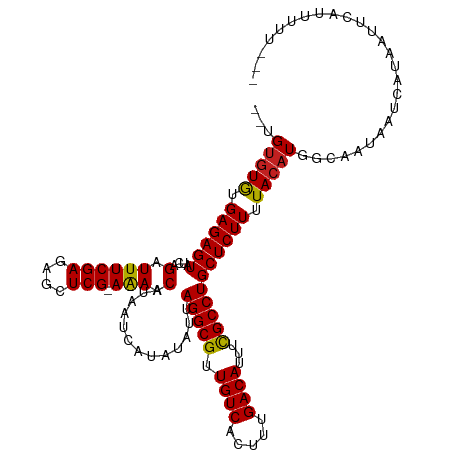
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:14 2011