| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,165,177 – 2,165,267 |
| Length | 90 |
| Max. P | 0.618119 |

| Location | 2,165,177 – 2,165,267 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.48 |
| Shannon entropy | 0.47450 |
| G+C content | 0.45566 |
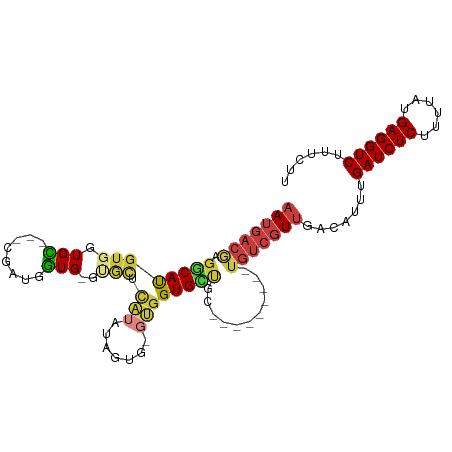
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -10.27 |
| Energy contribution | -11.05 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
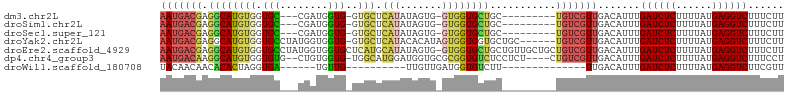
>dm3.chr2L 2165177 90 + 23011544 AAUGACGAGGCAUGUGGUGC---CGAUGGUG-GUGCUCAUAUAGUG-GUGGUGCUGC---------UGUCGUUGACAUUUGAUCUCUUUUAUGAGGUCUUUCUU ((((.((((((((...))))---)(((((..-(..(.(((......-))))..)..)---------)))).))).)))).((((((......))))))...... ( -30.10, z-score = -2.54, R) >droSim1.chr2L 2118861 90 + 22036055 AAUGACGAGGCAUGUGGUGC---CGAUGGUG-GUGCUCAUAUAGUG-GUGGUGCUGC---------UGUCGUUGACAUUUGAUCUCUUUUAUGAGGUCUUUCUU ((((.((((((((...))))---)(((((..-(..(.(((......-))))..)..)---------)))).))).)))).((((((......))))))...... ( -30.10, z-score = -2.54, R) >droSec1.super_121 33410 90 + 59987 AAUGACGAGGCAUGUGGUGC---CGAUGGUG-GUGCUCAUAUAGUG-GUGGUGCUGC---------UGUCGUUGACAUUUGAUCUCUUUUAUGAGGUCUUUCUU ((((.((((((((...))))---)(((((..-(..(.(((......-))))..)..)---------)))).))).)))).((((((......))))))...... ( -30.10, z-score = -2.54, R) >droYak2.chr2L 2136919 97 + 22324452 AAUGACGAGGCAUGUGGUGCCUAUGGUGGUG-GUGCUCAUACACAUAGUGGUGGUGCUGC------UGUCGUUGACAUUUGAUCUCUUUUAUGAGGUCUUUCUU ((((((.((((((...)))))).....((..-(..(.(((.(((...)))))))..)..)------))))))).......((((((......))))))...... ( -31.00, z-score = -1.69, R) >droEre2.scaffold_4929 2199898 103 + 26641161 AAUGACGAGGCAUGUGGUGCCUAUGGUGGUGCUCAUGCAUAUAGUG-GUGGUGCUGCUGUUGCUGCUGUCGUUGACAUUUGAUCUCUUUUAUGAGGUCUUUCUU ((((((.(((((((.((..((......))..)))))))...(((..-(..(.....)..)..))))))))))).......((((((......))))))...... ( -35.40, z-score = -2.66, R) >dp4.chr4_group3 7417863 97 - 11692001 AAUGACAAGGCAUGUGGUGUG--CUGUGGUG-UGGCAUGGAUGGUGCGCGGUGUCUCCUCU----CUGUCGUUGACAUUUGAUCUCUUUUAUGAGGUCUUUCCU (((((((.(((((.....)))--))(.((..-.(((((.(........).))))).)).).----.))))))).......((((((......))))))...... ( -26.30, z-score = -0.18, R) >droWil1.scaffold_180708 10437403 74 - 12563649 UACAACAACACACUAGGUGA------UGUUG----------UUGUUGAUGGUGUCUU--------------UUGACAUUUGAUCUCUUUUAUGAGGUCUUCGUU .((((((((((((...))).------)))))----------)))).(((((((((..--------------..)))))..((((((......))))))..)))) ( -24.20, z-score = -3.60, R) >consensus AAUGACGAGGCAUGUGGUGC___CGAUGGUG_GUGCUCAUAUAGUG_GUGGUGCUGC_________UGUCGUUGACAUUUGAUCUCUUUUAUGAGGUCUUUCUU (((((((.(((((............(((((....)).)))..........)))))...........))))))).......((((((......))))))...... (-10.27 = -11.05 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:23 2011