
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,545,012 – 1,545,111 |
| Length | 99 |
| Max. P | 0.996339 |

| Location | 1,545,012 – 1,545,111 |
|---|---|
| Length | 99 |
| Sequences | 14 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 66.89 |
| Shannon entropy | 0.71670 |
| G+C content | 0.37175 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -11.83 |
| Energy contribution | -12.06 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.996339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
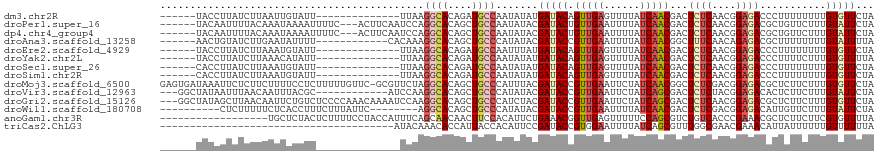
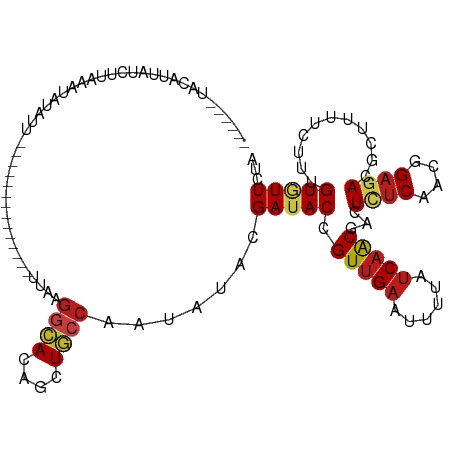
>dm3.chr2R 1545012 99 - 21146708 ------UACCUUAUCUUAAUUGUAUU--------------UUAAGGCACAGAUGCCAAUAUAUGAUACAGUUGAGUUUUAUCAACGACUCUCAACGGAGACCCUUUUUUUUGUGUUCUA ------........((((((((((((--------------....((((....)))).......)))))))))))).............((((....))))................... ( -18.00, z-score = -1.26, R) >droPer1.super_16 432276 110 - 1990440 ------UACAAUUUUACAAAUAAAAUUUUC---ACUUCAAUCCAGGCACAGCUGCCAAUAUACGAUACUGUUGAAUUUUAUCAACGACUCUCAACGGAGACGCUGUUCUUUGUAUUCUA ------........((((((..........---...((......((((....)))).............(((((......)))))))....((.((....)).))...))))))..... ( -16.70, z-score = -1.15, R) >dp4.chr4_group4 157273 110 + 6586962 ------UACAAUUUUACAAAUAAAAUUUUC---ACUUCAAUCCAGGCACAGCUGCCAAUAUACGAUACUGUUGAAUUUUAUCAACGACUCUCAACGGAGACGCUGUUCUUUGUAUUCUA ------........((((((..........---...((......((((....)))).............(((((......)))))))....((.((....)).))...))))))..... ( -16.70, z-score = -1.15, R) >droAna3.scaffold_13258 877754 101 + 1584923 ------AACUGUAUCUUGAAUAUUUU------------CACAAAGGCACAGCUGCCCAUAUACGAUACCGUUGAAUUUUAUCAACGGCUUUCAACAGAGACGCUUUUUUUUGUAUUUUA ------....((((..((((....))------------))....((((....))))...))))((..(((((((......)))))))...)).(((((((......)))))))...... ( -20.70, z-score = -2.55, R) >droEre2.scaffold_4929 5790266 99 + 26641161 ------UACCUUAUCUUAAAUGUAUU--------------UUAAGGCACAGAUGCCAAUUUAUGAUACAGUUGAGUUUUAUCAACGACUCUCAACGGAGACCCUUUUUUUUGUGUUCUA ------.......((((...((((((--------------.(((((((....))))...))).))))))((((((..((......))..)))))).))))................... ( -17.80, z-score = -1.19, R) >droYak2.chr2L 1285767 99 + 22324452 ------UACCUUAUCUUAAACAUAUU--------------UUAAGGCACAGAUGCCAAUAUAUGAUACAGUUGAGUUUUAUCAACGACUCUCAACGGAGACCCUUUUCUUUGUGUUUUA ------..............(((((.--------------....((((....))))...)))))((((((((((......)))))...((((....))))..........))))).... ( -17.80, z-score = -1.58, R) >droSec1.super_26 183042 99 - 826190 ------CACCUUAUCUUAAAUGUAUU--------------UUAAGGCACAGAUGCCAAUAUAUGAUACAGUUGAGUUUUAUCAACGACUCUCAACGGAGACCCUUUUUUUUGUGUUCUA ------.............(((((((--------------....((((....)))))))))))(((((((((((......)))))...((((....))))..........))))))... ( -18.20, z-score = -1.23, R) >droSim1.chr2R 437507 99 - 19596830 ------CACCUUAUCUUAAAUGUAUU--------------UUAAGGCACAGAUGCCAAUAUAUGAUACAGUUGAGUUUUAUCAACGACUCUCAACGGAGACCCUUUUUUUUGUGUUCUA ------.............(((((((--------------....((((....)))))))))))(((((((((((......)))))...((((....))))..........))))))... ( -18.20, z-score = -1.23, R) >droMoj3.scaffold_6500 12638942 118 - 32352404 GAGUGAUAAAUUCUCUUCUUUUCCUCUUUUUGUUC-GCGUUCUAGGCACAGCUGCCCAUUUACGAUACCGUUGAAUUCUAUCAACGGCUCUCGACGGAGACGCUCUUCUUUGUGUUCUA (((((......(((((((.................-.(((....((((....)))).....)))...(((((((......))))))).....)).)))))))))).............. ( -25.30, z-score = -0.76, R) >droVir3.scaffold_12963 2474509 104 - 20206255 ---GGCUAUAAUUUAACAAUUUACGC------------AUCCAAGGCACAGCUGCCCAUAUACGAUACCGUUGAAUUCUAUCAGCGACUCUCUACGGAGACACUCUUCUUUGUAUUCUA ---.((...((((....))))...))------------......((((....))))...((((((...((((((......))))))..((((....)))).........)))))).... ( -19.70, z-score = -1.64, R) >droGri2.scaffold_15126 5866670 116 + 8399593 ---GGCUAUAGCUUAACAAUUCUGUCUCCCCAAACAAAAUCCAAGGCACAGCUGCCCAUCUACGAUACCGUUGAAUUCUAUCAGCGACUCUCAACGGAGACGCUCUUCUUUGUGUUCUA ---(((....)))..((((...(((((((...............((((....))))............((((((......)))))).........))))))).......))))...... ( -22.70, z-score = -1.03, R) >droWil1.scaffold_180708 6728975 101 - 12563649 ----------CUCUUUUUCUCACCUUUCUUUAUUC--------AGGCACAGCUGCCCAUAUACGAUACCGUUGAAUUUUAUCAACGACUCUCGACGGAGACAUUGUUCUUUGUAUUCUA ----------.........................--------.((((....))))......(((...((((((......))))))....)))(((((((......)))))))...... ( -17.60, z-score = -1.76, R) >anoGam1.chr3R 24686635 101 - 53272125 ------------------UGCUCUACUCUUUUCCUACCAUUUCAGCAACAACUUCCACAUUCUGAAACGGUUGAGUUUUUCCAGCGUCUGUCACCCGAAACGCUCUUCUUCGUGUUUUA ------------------.(((..((((.......(((.((((((................)))))).))).))))......)))...........(((((((........))))))). ( -16.09, z-score = -1.23, R) >triCas2.ChLG3 25073499 80 - 32080666 ---------------------------------------AUACAAACACCAUUACCACAUUCCGAUACCGUGGAAUUUUAUCAGCGUUUGGCGAACGAAACAUUAUUUUUUGUUUUUUA ---------------------------------------...(((((.(.........((((((......)))))).......).)))))..((((((((.......)))))))).... ( -10.69, z-score = -0.59, R) >consensus ______UACAUUAUCUUAAAUAUAUU______________UUAAGGCACAGCUGCCAAUAUACGAUACCGUUGAAUUUUAUCAACGACUCUCAACGGAGACGCUUUUCUUUGUGUUCUA ............................................((((....)))).......(((((.(((((......)))))...((((....))))...........)))))... (-11.83 = -12.06 + 0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:07 2011