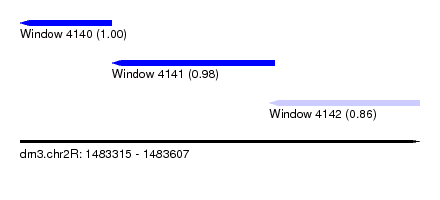
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,483,315 – 1,483,607 |
| Length | 292 |
| Max. P | 0.995450 |

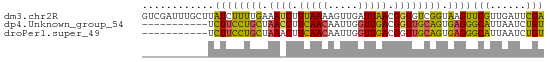
| Location | 1,483,315 – 1,483,382 |
|---|---|
| Length | 67 |
| Sequences | 3 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 62.63 |
| Shannon entropy | 0.48966 |
| G+C content | 0.40609 |
| Mean single sequence MFE | -16.40 |
| Consensus MFE | -11.48 |
| Energy contribution | -9.93 |
| Covariance contribution | -1.54 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.995450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1483315 67 - 21146708 GUCGAUUUGCUUAUCUUUUGAAAUCUUUAAAAGUUGAUUAACGGUGUCGGUAAGUUCGUUGAUUCGA ((((((..(((((((((((((.....))))))...(((.......))))))))))..)))))).... ( -12.70, z-score = -0.82, R) >dp4.Unknown_group_54 46636 56 - 61718 -----------UCUUCCUGCUAACCUUCAACAAUUGGUUGACGGUUGCAGUGAGGGCAUUAAUCUGU -----------(((((((((.((((.(((((.....))))).)))))))).)))))........... ( -20.70, z-score = -3.71, R) >droPer1.super_49 18114 56 + 569410 -----------UCUUCCUGCUAAACUUCAACAAUUGGUUGACGGUUGCAGUGAGGGCAUUAAUCUGU -----------(((((((((..(((((((((.....))))).)))))))).)))))........... ( -15.80, z-score = -2.08, R) >consensus ___________UCUUCCUGCUAAACUUCAACAAUUGGUUGACGGUUGCAGUGAGGGCAUUAAUCUGU ............((((((((.((((.(((((.....))))).)))))))).))))(((......))) (-11.48 = -9.93 + -1.54)
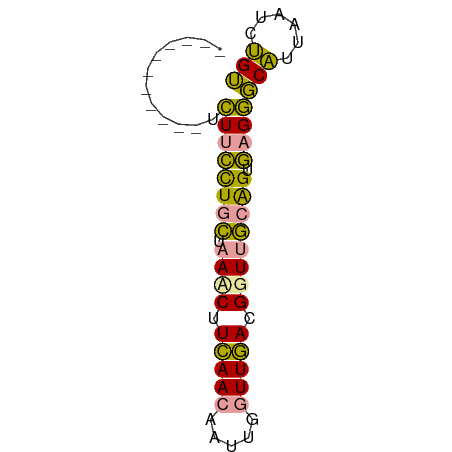

| Location | 1,483,382 – 1,483,501 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 54.08 |
| Shannon entropy | 0.58159 |
| G+C content | 0.36695 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -11.86 |
| Energy contribution | -11.43 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1483382 119 - 21146708 GUCCAGAAAGACUGGACUAACUACGUUAAUUUUUGCGAGUGUGAUUGUAAGCAAAAGGUUCUAUGUUCCAUG-ACAAUGAUUCUAUUGCGUGUUAAUAGCAUUUCGUAAAGGUGGCCUGU ((((((.....)))))).......((((.(((((((((((((.((((...((((.((..((..((((....)-)))..))..)).))))....)))).))).)))))))))))))).... ( -32.10, z-score = -2.07, R) >dp4.Unknown_group_54 46692 91 - 61718 AUCAAUGAUAGACGGUUUGAUGGUGUUUAUUUUCGCCAAAGUGGUGGCUGUCAUUAUGUUUGGUAAUUCAAGCGUCAGAAUUAUAUUUCUU----------------------------- ...((((((((.((.(....(((((........)))))....).)).))))))))((((((((....)))))))).((((......)))).----------------------------- ( -21.80, z-score = -1.92, R) >droPer1.super_49 18170 91 + 569410 AUCAAUGAUAGACGGUUUGAUGGUGUUUAUUUGCGCCAAAGUGGUGGCUGUCAUUAUGUUUGGUAAUUCAAGCGUCAGAAUUAUAUUUCUU----------------------------- ...((((((((.((.(....((((((......))))))....).)).))))))))((((((((....)))))))).((((......)))).----------------------------- ( -23.80, z-score = -2.15, R) >consensus AUCAAUGAUAGACGGUUUGAUGGUGUUUAUUUUCGCCAAAGUGGUGGCUGUCAUUAUGUUUGGUAAUUCAAGCGUCAGAAUUAUAUUUCUU_____________________________ ........((((((...(((((((..((((((......))))))..)))))))...))))))(((((((........))))))).................................... (-11.86 = -11.43 + -0.42)

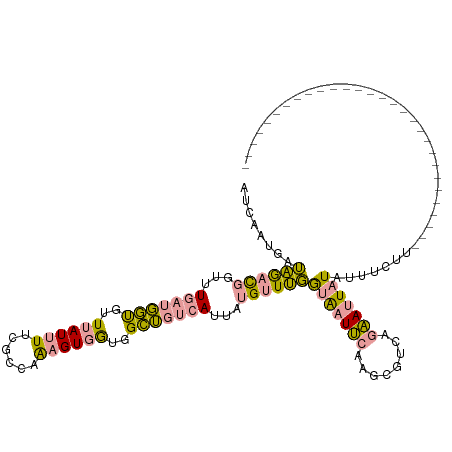
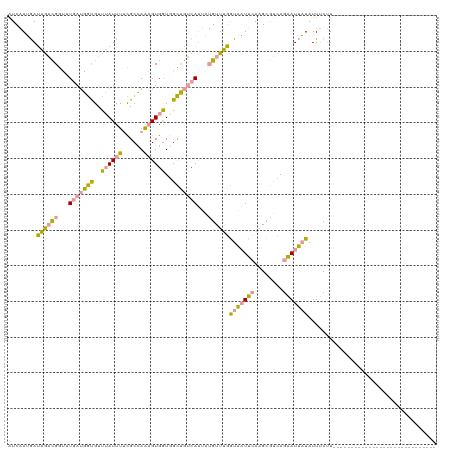
| Location | 1,483,497 – 1,483,607 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 52.60 |
| Shannon entropy | 0.66773 |
| G+C content | 0.39877 |
| Mean single sequence MFE | -21.61 |
| Consensus MFE | -9.69 |
| Energy contribution | -7.71 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1483497 110 - 21146708 GUAAAACGUUAAGGGAUUGUAAAACCUUUACGGACGCAACGGACAAAAUUUCCCUUAUGGGGGUGUUGGUGUGCUCCUCGCCCCAAACAUUCUCCAAGUCUGUAUGGUCC .......((.((((..........)))).))((((.((((((((.........(....)((((((..((......)).)))))).............)))))).)))))) ( -30.80, z-score = -0.71, R) >dp4.Unknown_group_54 46779 90 - 61718 --------------------AAGGCACUCACAUUGGUAGCGUUCAUUAAUUGGAAUACGCUGAUACCGGUAGAACUUUAAUUCAAAAUCUCUUCCAAAUCAUCGCUAUCA --------------------.............((((((((........((((((...((((....)))).(((......)))........)))))).....)))))))) ( -17.12, z-score = -1.70, R) >droPer1.super_49 18257 107 + 569410 -UUAAUUCUUUAUUAAC--UAAGGCACUCACAUUGGUAGCGUUCAUUAAUUGGGAUACGCUGAUACCGGUAGAACUUUUAUUCAAAAUCUCUUCCAAAUCAUCGCUAUCA -......(((((.....--))))).........((((((((..........(((((..((((....)))).(((......)))...)))))...........)))))))) ( -16.90, z-score = -1.13, R) >consensus _U_AA___UU_A___A___UAAGGCACUCACAUUGGUAGCGUUCAUUAAUUGGAAUACGCUGAUACCGGUAGAACUUUAAUUCAAAAUCUCUUCCAAAUCAUCGCUAUCA ..................................(((((((.......(((((......)))))...((.(((..............)))...)).......))))))). ( -9.69 = -7.71 + -1.99)

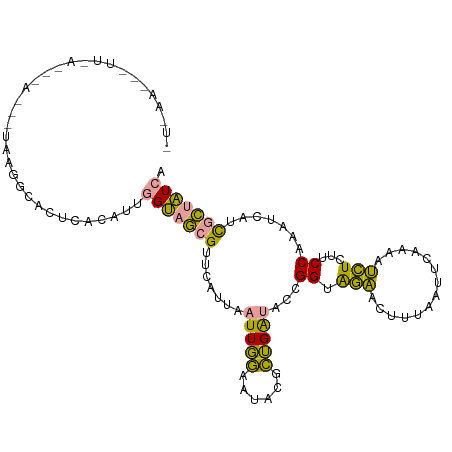
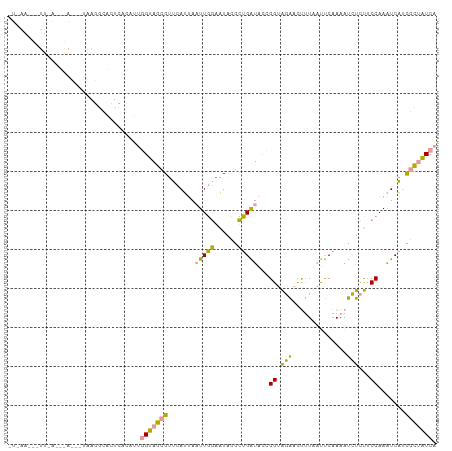
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:06 2011