| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,343,197 – 1,343,307 |
| Length | 110 |
| Max. P | 0.913272 |

| Location | 1,343,197 – 1,343,307 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 57.19 |
| Shannon entropy | 0.59260 |
| G+C content | 0.30686 |
| Mean single sequence MFE | -14.52 |
| Consensus MFE | -7.43 |
| Energy contribution | -6.77 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913272 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
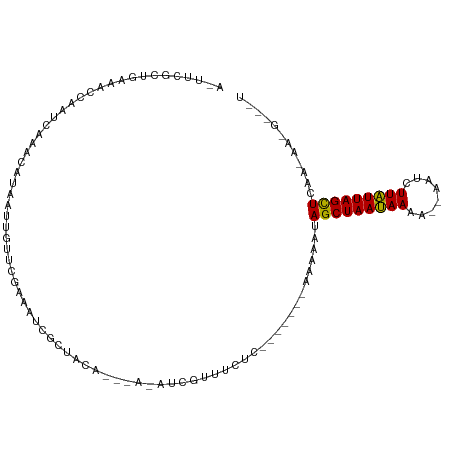
>dm3.chr2R 1343197 110 - 21146708 AUUUCGCUGAAACCAGUUAAACAGAAUCGUUCGAAAUCGCUACAAAAAAAUCGUCUUUCUUUUAUCAAAAAUAGCUAAAAAAGAUUUUUUUUUUAGCUCAAAAACGACGU .(((.((((....)))).))).....(((((.......((((.(((((((..((((((..((((.(.......).))))))))))))))))).)))).....)))))... ( -15.00, z-score = -0.08, R) >droEre2.scaffold_4929 22653860 91 - 26641161 --UUCGCUGAGCCCAAUC-AACGUAAUUGUUCAGAAUCGCUUU-------UUGUUGCUC-------AAAAAUAGCUAAUAAAA--AAGCUUAUUAGUUCAACAAUGGUAU --....((((((......-.........))))))....(((..-------((((((...-------......(((((((((..--....))))))))))))))).))).. ( -15.96, z-score = -1.11, R) >droYak2.chr2L_random 108378 90 - 4064425 AGUUCGAACCAACAAAUGAAACAUAAUUAUUUGAAAUCGCUACA--CACAUCAUUUCUC------AAGAUACAGCUAAUAAAA--CAUCUUAUUAGCUCA---------- .((.(((.....(((((((.......)))))))...)))..)).--.............------.......(((((((((..--....)))))))))..---------- ( -12.60, z-score = -2.93, R) >consensus A_UUCGCUGAAACCAAUCAAACAUAAUUGUUCGAAAUCGCUACA___A_AUCGUUUCUC_______AAAAAUAGCUAAUAAAA__AAUCUUAUUAGCUCAA_AA_G___U ........................................................................(((((((((........)))))))))............ ( -7.43 = -6.77 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:02:00 2011