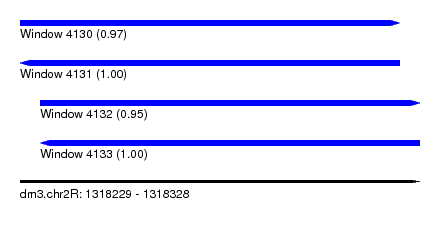
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,318,229 – 1,318,328 |
| Length | 99 |
| Max. P | 0.999764 |

| Location | 1,318,229 – 1,318,323 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 59.03 |
| Shannon entropy | 0.65374 |
| G+C content | 0.43636 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -9.18 |
| Energy contribution | -10.05 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1318229 94 + 21146708 GCGCUUUGGAGAUUCAUCUAAUAGCUUAAGGUCAUUAAACUGUGAAUUAAUCGCGGAAAGAAGAUCAUCGGCUCGACGAAAACUAUCUCUAGCU ..(((..((((((...((....((((...((((.((...((((((.....))))))...)).))))...))))....)).....))))))))). ( -20.90, z-score = -0.52, R) >dp4.Unknown_group_58 28543 85 - 69560 GCACCUCGGAGACUCAUUCAGCAGUUUUAAACCGCUAAACUGAGCUUCUAUGGCUAGGAGCCGAAGGUAUUGUUUUUUAAAACCA--------- ..(((((((((.((((((.(((.((.....)).))).)).))))))))...(((.....)))).)))).................--------- ( -19.70, z-score = -0.60, R) >droPer1.super_2 225673 85 + 9036312 GCGCCUCGGAGACUCAUUCACCAGUUUUAAACAGCUAAACUGAGCCUCCAUGGCUAGGAGCCGAUCGUAUUGGUUUUUAAAACCA--------- ..(((..((((.((((......((((......))))....)))).))))..)))(((((((((((...)))))))))))......--------- ( -24.50, z-score = -2.32, R) >droWil1.scaffold_181154 2082159 85 + 4610121 ACGUUUCGGAGACUCAUUCAUAAGCGAUAAACUUCUGAACUGAGUCUCCACCGCAGUAAACUCCGCUUGGAGUUUGUUAAAACCU--------- ..((((.(((((((((((((.(((.......))).)))).))))))))).......((((((((....))))))))...))))..--------- ( -29.20, z-score = -4.66, R) >consensus GCGCCUCGGAGACUCAUUCAACAGCUUUAAACCGCUAAACUGAGCCUCCAUCGCUAGAAGCCGAUCGUAGUGUUUUUUAAAACCA_________ ..(((..((((.((((((((((.((.....)).)))))).)))).))))..)))........................................ ( -9.18 = -10.05 + 0.87)


| Location | 1,318,229 – 1,318,323 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 59.03 |
| Shannon entropy | 0.65374 |
| G+C content | 0.43636 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -17.52 |
| Energy contribution | -18.90 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1318229 94 - 21146708 AGCUAGAGAUAGUUUUCGUCGAGCCGAUGAUCUUCUUUCCGCGAUUAAUUCACAGUUUAAUGACCUUAAGCUAUUAGAUGAAUCUCCAAAGCGC .(((.(((((.....((((((...))))))((.(((....(.((.....)).)(((((((.....)))))))...))).)))))))...))).. ( -17.90, z-score = 0.10, R) >dp4.Unknown_group_58 28543 85 + 69560 ---------UGGUUUUAAAAAACAAUACCUUCGGCUCCUAGCCAUAGAAGCUCAGUUUAGCGGUUUAAAACUGCUGAAUGAGUCUCCGAGGUGC ---------..((((....))))..((((((.(((.....)))...((..((((.(((((((((.....)))))))))))))..)).)))))). ( -28.30, z-score = -3.34, R) >droPer1.super_2 225673 85 - 9036312 ---------UGGUUUUAAAAACCAAUACGAUCGGCUCCUAGCCAUGGAGGCUCAGUUUAGCUGUUUAAAACUGGUGAAUGAGUCUCCGAGGCGC ---------(((((.....)))))................(((.((((((((((.((((.(.((.....)).).)))))))))))))).))).. ( -28.80, z-score = -2.73, R) >droWil1.scaffold_181154 2082159 85 - 4610121 ---------AGGUUUUAACAAACUCCAAGCGGAGUUUACUGCGGUGGAGACUCAGUUCAGAAGUUUAUCGCUUAUGAAUGAGUCUCCGAAACGU ---------..........(((((((....)))))))...(((.((((((((((.((((.((((.....)))).))))))))))))))...))) ( -33.00, z-score = -4.59, R) >consensus _________UGGUUUUAAAAAACAAUACGAUCGGCUCCCAGCCAUGGAGGCUCAGUUUAGCGGUUUAAAACUGAUGAAUGAGUCUCCGAAGCGC ........................................(((.((((((((((.(((((((((.....))))))))))))))))))).))).. (-17.52 = -18.90 + 1.38)

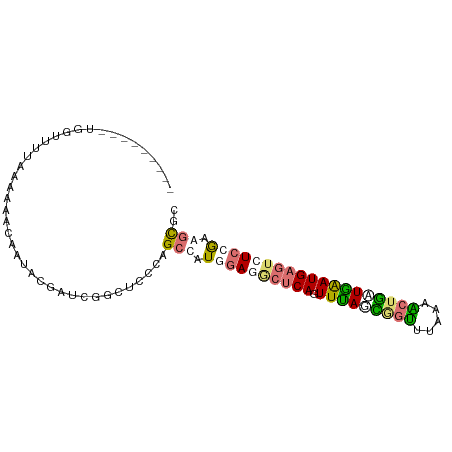
| Location | 1,318,234 – 1,318,328 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 57.47 |
| Shannon entropy | 0.66036 |
| G+C content | 0.42294 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -7.71 |
| Energy contribution | -8.03 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1318234 94 + 21146708 UUGGAGAUUCAUCUAAUAGCUUAAGGUCAUUAAACUGUGAAUUAAUCGCGGAAAGAAGAUCAUCGGCUCGACGAAAACUAUCUCUAGCUACGCC ((((((((...((....((((...((((.((...((((((.....))))))...)).))))...))))....)).....))))))))....... ( -19.30, z-score = -0.39, R) >dp4.Unknown_group_58 28548 80 - 69560 UCGGAGACUCAUUCAGCAGUUUUAAACCGCUAAACUGAGCUUCUAUGGCUAGGAGCCGAAGGUAUUGUUUUUUAAAACCA-------------- ..((((.((((((.(((.((.....)).))).)).))))))))..((((.....))))..(((.(((.....))).))).-------------- ( -19.00, z-score = -0.87, R) >droPer1.super_2 225678 80 + 9036312 UCGGAGACUCAUUCACCAGUUUUAAACAGCUAAACUGAGCCUCCAUGGCUAGGAGCCGAUCGUAUUGGUUUUUAAAACCA-------------- ..((((.((((......((((......))))....)))).)))).(((.(((((((((((...)))))))))))...)))-------------- ( -20.50, z-score = -1.79, R) >droWil1.scaffold_181154 2082164 80 + 4610121 UCGGAGACUCAUUCAUAAGCGAUAAACUUCUGAACUGAGUCUCCACCGCAGUAAACUCCGCUUGGAGUUUGUUAAAACCU-------------- ..(((((((((((((.(((.......))).)))).))))))))).......((((((((....)))))))).........-------------- ( -28.80, z-score = -5.22, R) >consensus UCGGAGACUCAUUCAACAGCUUUAAACCGCUAAACUGAGCCUCCAUCGCUAGAAGCCGAUCGUAGUGUUUUUUAAAACCA______________ ..((((.((((((((((.((.....)).)))))).)))).)))).................................................. ( -7.71 = -8.03 + 0.31)

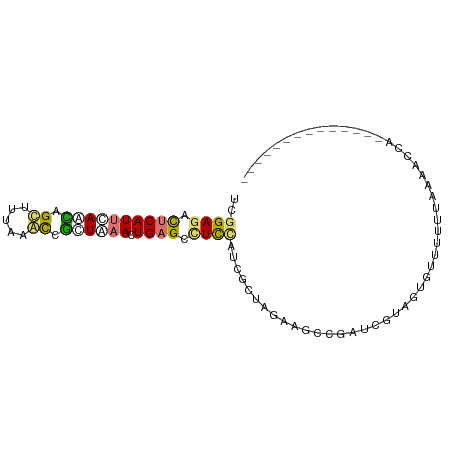
| Location | 1,318,234 – 1,318,328 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 57.47 |
| Shannon entropy | 0.66036 |
| G+C content | 0.42294 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -15.25 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.59 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.94 |
| SVM RNA-class probability | 0.999488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1318234 94 - 21146708 GGCGUAGCUAGAGAUAGUUUUCGUCGAGCCGAUGAUCUUCUUUCCGCGAUUAAUUCACAGUUUAAUGACCUUAAGCUAUUAGAUGAAUCUCCAA ((....((.(((((.((...((((((...)))))).)))))))..))((((...((..(((((((.....)))))))....))..)))).)).. ( -16.80, z-score = 0.65, R) >dp4.Unknown_group_58 28548 80 + 69560 --------------UGGUUUUAAAAAACAAUACCUUCGGCUCCUAGCCAUAGAAGCUCAGUUUAGCGGUUUAAAACUGCUGAAUGAGUCUCCGA --------------.(((.............)))...(((.....)))...((..((((.(((((((((.....)))))))))))))..))... ( -23.12, z-score = -2.70, R) >droPer1.super_2 225678 80 - 9036312 --------------UGGUUUUAAAAACCAAUACGAUCGGCUCCUAGCCAUGGAGGCUCAGUUUAGCUGUUUAAAACUGGUGAAUGAGUCUCCGA --------------(((((.....)))))........(((.....))).((((((((((.((((.(.((.....)).).)))))))))))))). ( -26.70, z-score = -3.13, R) >droWil1.scaffold_181154 2082164 80 - 4610121 --------------AGGUUUUAACAAACUCCAAGCGGAGUUUACUGCGGUGGAGACUCAGUUCAGAAGUUUAUCGCUUAUGAAUGAGUCUCCGA --------------..........(((((((....))))))).......((((((((((.((((.((((.....)))).)))))))))))))). ( -30.10, z-score = -3.98, R) >consensus ______________UGGUUUUAAAAAACAAUACGAUCGGCUCCCAGCCAUGGAGGCUCAGUUUAGCGGUUUAAAACUGAUGAAUGAGUCUCCGA .....................................(((.....))).((((((((((.(((((((((.....))))))))))))))))))). (-15.25 = -16.12 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:59 2011