| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,143,460 – 1,143,525 |
| Length | 65 |
| Max. P | 0.557599 |

| Location | 1,143,460 – 1,143,525 |
|---|---|
| Length | 65 |
| Sequences | 5 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 69.53 |
| Shannon entropy | 0.53175 |
| G+C content | 0.41791 |
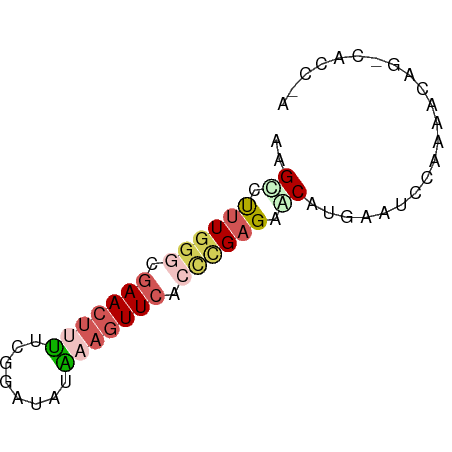
| Mean single sequence MFE | -12.86 |
| Consensus MFE | -6.58 |
| Energy contribution | -7.94 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.557599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1143460 65 - 21146708 AAGCCUUU-GGCGAACUUUUCGGACAAAAUGUUCACCAGAGAGCAUAAAUCCAAUACAGUCACCGA ..((((((-((.((((((((.....)))).)))).)))))).))...................... ( -15.60, z-score = -2.60, R) >droSec1.super_59 75263 66 - 165940 AAGCCUUUGGGAGAACUUUUCGGAUAUAAAGUUCAAUCGAGAACGUGAAUCCAAAACAGGCACCAA ..(((((((((.(((((((........)))))))..(((......))).)))))...))))..... ( -16.10, z-score = -1.88, R) >droSim1.chr2R 111651 66 - 19596830 AAGCCUUUGGGAGAACUUUUCGGAUAUAAAGUUCAAUCGAGAACGUAAAUCCAAAACAGGCACCAA ..(((((((((.(((((((........)))))))...((....))....)))))...))))..... ( -15.40, z-score = -2.13, R) >droMoj3.scaffold_6489 31253 59 + 80705 UAGUGCUUGUGCGAACUGUUCUGAGAUGAAGUUCACCUGUGAGCAAGAAUCCAAUAGAG------- ...((((..((.(((((..((......))))))).)..)..))))..............------- ( -10.50, z-score = 0.01, R) >droYak2.chrU 19290094 53 + 28119190 AAGUCUUUGGGCAAACUCCUCGGAUAUGAAAUUCACCUGAGAACAUGAAUCCA------------- ..(((....)))......(((((...((.....)).)))))............------------- ( -6.70, z-score = 0.99, R) >consensus AAGCCUUUGGGCGAACUUUUCGGAUAUAAAGUUCACCCGAGAACAUGAAUCCAAAACAG_CACC_A ..((.((((((.(((((((........))))))).)))))).))...................... ( -6.58 = -7.94 + 1.36)

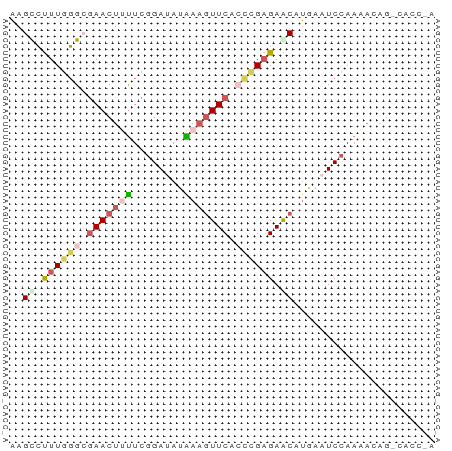
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:49 2011