
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,092,307 – 1,092,369 |
| Length | 62 |
| Max. P | 0.500000 |

| Location | 1,092,307 – 1,092,369 |
|---|---|
| Length | 62 |
| Sequences | 5 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 51.01 |
| Shannon entropy | 0.90252 |
| G+C content | 0.47794 |
| Mean single sequence MFE | -13.08 |
| Consensus MFE | -5.38 |
| Energy contribution | -5.22 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.28 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
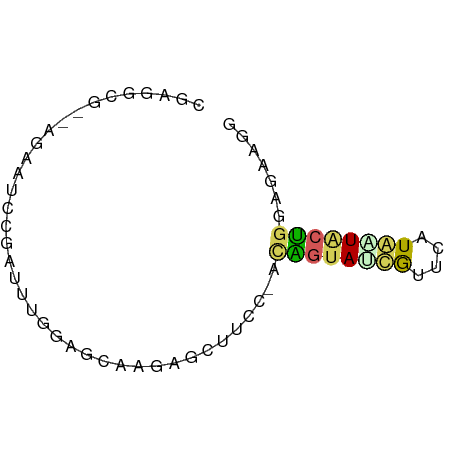
>dm3.chr2R 1092307 62 + 21146708 CGAGGCG--AGAAUACGAUUUGGAGCCAAAGUUUUC-ACAGUACCGUUUCUAAUACUGGAGAAGG ...(((.--.((((...))))...)))....(((((-.(((((..........)))))))))).. ( -11.40, z-score = -0.22, R) >droSec1.super_59 38456 62 + 165940 CGAGGCG--AGAAUCCGAUUUGGAGGAAAAGCAUCC-ACAGUAUUGUCCAUAGUUCUGGAGAAGG .((.((.--....(((.....)))......)).)).-......((.((((......)))).)).. ( -10.00, z-score = 0.93, R) >droSim1.chr2h_random 2333780 64 - 3178526 CGGUUCGGAAGAAUCCGGUUUAGAGGAAGGUCAUUC-ACAAUAUCGUUCAUGAUACUGGAGAAGG ((((((....))).)))................(((-.((.(((((....))))).))))).... ( -12.70, z-score = -0.08, R) >droYak2.chrU 21364062 58 - 28119190 -----CGGUGGGAA--AGUUUACAACCGGAAUUUCGGAGGGUACCAUUCAUGGUACUGGCACACG -----..(((....--.........((((....))))..(((((((....)))))))....))). ( -15.20, z-score = -1.12, R) >droPer1.super_330 45385 62 + 52110 UGAAACGAAGGUGUGCUUUGGGAACAAGGAGAUCCCAAUGGUAUUACUCGUAUUGCCGGGUG--- ....((....))(((((((((((.(.....).)))))).)))))((((((......))))))--- ( -16.10, z-score = -0.89, R) >consensus CGAGGCG__AGAAUCCGAUUUGGAGCAAGAGCUUCC_ACAGUAUCGUUCAUAAUACUGGAGAAGG ......................................((((((((....))))))))....... ( -5.38 = -5.22 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:48 2011