
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 1,091,313 – 1,091,403 |
| Length | 90 |
| Max. P | 0.670001 |

| Location | 1,091,313 – 1,091,403 |
|---|---|
| Length | 90 |
| Sequences | 13 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 76.81 |
| Shannon entropy | 0.52334 |
| G+C content | 0.34493 |
| Mean single sequence MFE | -18.05 |
| Consensus MFE | -9.98 |
| Energy contribution | -9.13 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 1091313 90 + 21146708 AGACGUUCUCUGUUGAUUUGGAUUA-ACGUUUAUAGUGUAUAUAUCUAGAGCCUCCAUCGCAUAGUAAACCAAGUCGAUGUAAGAACAUAC ....(((((..((((((((((.(((-.....((((.....)))).(((..((.......)).)))))).))))))))))...))))).... ( -17.90, z-score = -0.81, R) >droYak2.chr2L_random 2877525 90 + 4064425 AGACGUUCUCUGUUGAUUUGGAUUC-ACAUUUAUAGUGUAUAUAUCUAAAGCCUCCAUCGCAUAGUGAACCAAGUCGAUGUAGGAACAUAC ....(((((..((((((((((.(((-((...((((.....))))......((.......))...)))))))))))))))...))))).... ( -23.80, z-score = -2.52, R) >droAna3.scaffold_13266 19823855 90 + 19884421 AGACGUUCUUUGUUGGCUUGGAUUG-ACAUUAAUGGUAUAAAUAUCCAAAGCCUCCAUUGCAAAAUAAACCAAGUCAAUGUAAGAACAUAA ....(((((((((((((((((.(((-....((((((..................)))))).....))).))))))))))).)))))).... ( -23.77, z-score = -3.42, R) >droPer1.super_43 279566 90 + 623856 UGAAGUUCGCUGUUGGUUUGGAUUA-ACGUUAAUUGUAUAAAUAUCUAAAGCCUCCAUUGCAAAAUGAACUAAGUCAAUAUAUACACAUAA ((((((((((((..(((((((((..-(((.....)))......)))).)))))..))..)).....)))))...))).............. ( -10.50, z-score = 0.93, R) >dp4.Unknown_group_316 11887 90 + 15256 UGAAGUUCGCUGUUGGUUUGGAUUA-ACGUUAAUUGUAUAAAUAUCUAAAGCCUCCAUUGCAAAAUGAACUAAGUCAAUAUAUACACAUAA ((((((((((((..(((((((((..-(((.....)))......)))).)))))..))..)).....)))))...))).............. ( -10.50, z-score = 0.93, R) >droSim1.chr2R 67694 90 + 19596830 AGAAGUUCUCUGUUGAUUUGGAUUA-ACGUUUAUAGUGUAUAUAUCUAAAGCCUCCAUCGCAUAGUAAACCAAGUCGAUGUAAGAACAUAC ....(((((..((((((((((.((.-((...((((.....))))......((.......))...)).))))))))))))...))))).... ( -17.50, z-score = -1.41, R) >droSec1.super_59 37251 90 + 165940 AGAAGUUCUCUGUUGAUUUGGAUUA-ACGUUUAUAGUGUAUAUAUCUAAAGCCUCCAUCGCAUAGUAAACCAAGUCGAUAUAAGAACAUAC ....(((((.(((((((((((.((.-((...((((.....))))......((.......))...)).)))))))))))))..))))).... ( -18.20, z-score = -1.90, R) >droEre2.scaffold_4929 5115488 90 - 26641161 AGACGUUCUCUGUUGAUUUGGAUUA-ACGUUUAUAGUGUAUAUAUCUAAAGCCUCCAUCGCAUAGUGAACCAAGUCGAUGUAAGAACAUAC ....(((((..((((((((((....-..((((.(((.........))))))).....((((...)))).))))))))))...))))).... ( -19.60, z-score = -1.47, R) >droGri2.scaffold_15245 11954460 90 + 18325388 AGAUGAGCGCUGUUGAUUUGGGUUA-ACAUUUAUUGUAUAUAUAUCCAAAGCUUCCAUUGCAUAAUGCACUAGAUCAAUAUAUGUGCAUAA ......(((((((((((((((....-(((.....))).....................(((.....))))))))))))))...)))).... ( -18.80, z-score = -0.28, R) >droMoj3.scaffold_6496 12130334 90 - 26866924 AGAUGCUCGUUGUUGAUUAGGAUUU-ACAUUAAUGGUAUAUAUAUCUAAAGCUUCCAUUGCAAAAUGCACCAAGUCAAUGUAGGUACACAA ...((((...((((((((.((....-.....(((((..................)))))((.....)).)).))))))))..))))..... ( -14.07, z-score = 0.74, R) >droVir3.scaffold_12875 12082855 90 + 20611582 AGAUGCUCGUUGUUGAUUUGGGUUA-ACGUUUAUUGUAUAUAUAUCUAACGCUUCCAUUGCAAAAUGCACCAAGUCAAUAUAUAUGCAUAA ..((((....(((((((((((....-.((((...(((....)))...)))).......(((.....)))))))))))))).....)))).. ( -21.90, z-score = -2.20, R) >droWil1.scaffold_180700 1506162 90 - 6630534 AGAUGUACGCUGUUGGUUUGGAUUC-ACAUUUAUUGUAUAUAUAUCCAAAGCCUCCAUUGCAAAAUGAACCAACUCAAUAUACGUGCAAAG ...((((((..((((((((((((..-(((.....)))......))))).......((((....)))))))))))........))))))... ( -18.50, z-score = -1.47, R) >triCas2.ChLG6 12688674 84 - 13544221 UUUGGGGACCUGUCACUUGAGGUCCCGGAUACAACGUAUAGAUAUCUAACGCUUUCAAAGCCCACUUAACCAAUCUGACAAAAA------- (((((((((((........)))))))((.......((.(((....)))))(((.....)))........)).......))))..------- ( -19.60, z-score = -1.63, R) >consensus AGAAGUUCGCUGUUGAUUUGGAUUA_ACGUUUAUAGUAUAUAUAUCUAAAGCCUCCAUUGCAAAAUGAACCAAGUCAAUAUAAGAACAUAA ..........(((((((((((.....((.......)).............((.......))........)))))))))))........... ( -9.98 = -9.13 + -0.85)

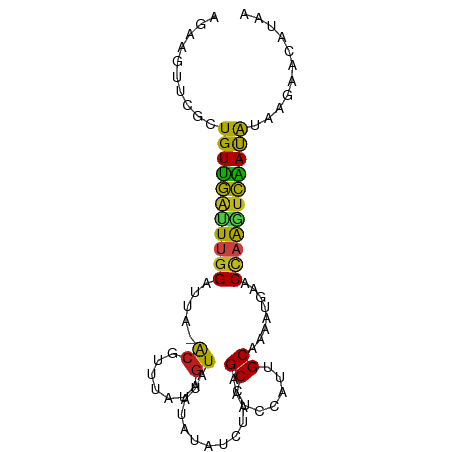
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:47 2011