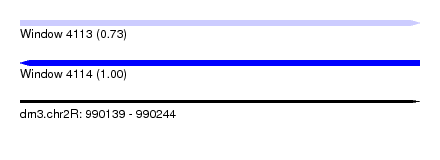
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 990,139 – 990,244 |
| Length | 105 |
| Max. P | 0.997142 |

| Location | 990,139 – 990,244 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.21 |
| Shannon entropy | 0.48164 |
| G+C content | 0.55082 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -22.10 |
| Energy contribution | -21.27 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
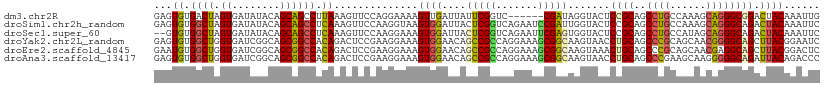
>dm3.chr2R 990139 105 + 21146708 GAGUGUGACUAGUGAUAUACAGCAGCCUUAAAGUUCCAGGAAAAGUUGAUUAUUCGGUC------CGAUAGGUACUCCGCAGCCUGCCAAAGCAGGGCGGACUACAAAUUG ......((((((((((...((((..(((.........)))....)))))))))).))))------((((..(((.(((((..(((((....)))))))))).)))..)))) ( -28.70, z-score = -1.06, R) >droSim1.chr2h_random 1002769 111 + 3178526 GAGUGUGGCUAGUGAUAUACAGCAGCCUCAAAGUUCCAAGGUAAGUGGAUUACUCGGUCAGAAUCCGAUUGGUACUCCGCAGCCUGCCAAAGCAGGGCAGACUACAAAUUC (((.((.(((.(((...)))))).)))))..((((.........(((((.(((.(((((.......)))))))).)))))..(((((....)))))...))))........ ( -31.20, z-score = -0.65, R) >droSec1.super_60 143709 109 - 163194 --GUGUGGCUAGUGAUAUACAGCAGCCUCAAAGUUCCAAGGAAAGUGGAUUACUCGGUCAGAAUUCGAGUGGUACUCCGCAGCCUGCCAUAGCAGGGCAGACUACAAAUUC --.((.((((.((........)))))).))...(((....))).(((((((((((((.......))))))))...)))))((.(((((.......))))).))........ ( -32.00, z-score = -1.00, R) >droYak2.chr2L_random 1913381 111 + 4064425 GAGUGUGGCUGGUGAUCGGCAGCGGCCACAGACUCCGAAGGAAAGUGGAACAGCCGCCAGGAAAGCGGCAAGUAACCUGCAGCCCGCAGCAACGGGGCAGCUUACGGAAUC ...((((((((.((.....)).))))))))((.(((................(((((.......)))))..((((.((((..((((......)))))))).))))))).)) ( -44.10, z-score = -1.51, R) >droEre2.scaffold_4845 974651 111 + 22589142 GAAUGUGGCUGGUGAUCGGCAGCGGCCACAGACUCCGAAGGAAAGUGGAACAGCCGCCAGGAAAGCGGCAAGUAAACUGCAGCCCGCAGCAACGAGGCAGCUUACGGACUC ...((((((((.((.....)).))))))))((.(((((((....(..(.((.(((((.......)))))..))...)..).(((((......)).)))..))).)))).)) ( -40.00, z-score = -1.01, R) >droAna3.scaffold_13417 6855903 111 + 6960332 GAGUGUGGCUGGUGAUCGGCAGCGGCCACAGACUCCGAAGGAAAGUGGAACAGCCGCCAGGAAAGCGGCAAGUAACCUGCAGCCCGAAGCAAGGGGGCAGAUUACAGACCC (((((((((((.((.....)).)))))))..))))....((...........(((((.......)))))..((((.((((..(((.......))).)))).))))...)). ( -43.20, z-score = -2.07, R) >consensus GAGUGUGGCUAGUGAUAGACAGCAGCCACAAACUCCCAAGGAAAGUGGAACAGCCGCCAGGAAAGCGACAAGUAAUCCGCAGCCCGCAACAACAGGGCAGACUACAAAUUC ...((.((((.((........)))))).))..............((((....(((((.......))))).......((((..((((......)))))))).))))...... (-22.10 = -21.27 + -0.83)
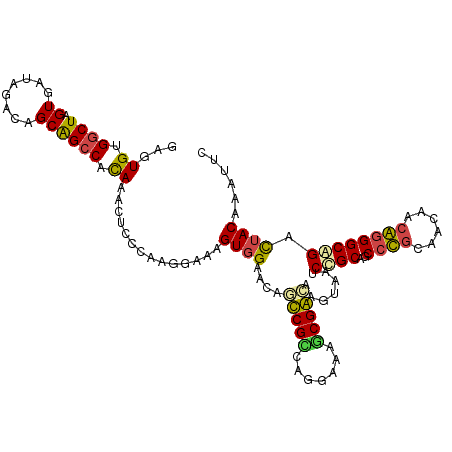

| Location | 990,139 – 990,244 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.21 |
| Shannon entropy | 0.48164 |
| G+C content | 0.55082 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -28.31 |
| Energy contribution | -25.90 |
| Covariance contribution | -2.41 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.997142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
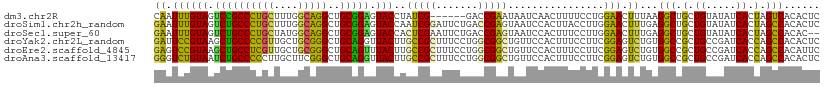
>dm3.chr2R 990139 105 - 21146708 CAAUUUGUAGUCCGCCCUGCUUUGGCAGGCUGCGGAGUACCUAUCG------GACCGAAUAAUCAACUUUUCCUGGAACUUUAAGGCUGCUGUAUAUCACUAGUCACACUC ......(((.((((((((((....)))))..))))).))).....(------..(((...((......))...)))..).....(((((.((.....)).)))))...... ( -26.40, z-score = -0.76, R) >droSim1.chr2h_random 1002769 111 - 3178526 GAAUUUGUAGUCUGCCCUGCUUUGGCAGGCUGCGGAGUACCAAUCGGAUUCUGACCGAGUAAUCCACUUACCUUGGAACUUUGAGGCUGCUGUAUAUCACUAGCCACACUC ......((((((((((.......))))))))))(((((.((((((((.......))))((((.....)))).)))).)))))..(((((.((.....)).)))))...... ( -36.50, z-score = -2.52, R) >droSec1.super_60 143709 109 + 163194 GAAUUUGUAGUCUGCCCUGCUAUGGCAGGCUGCGGAGUACCACUCGAAUUCUGACCGAGUAAUCCACUUUCCUUGGAACUUUGAGGCUGCUGUAUAUCACUAGCCACAC-- ((((((((((((((((.......)))))))))).(((.....)))))))))...(((((.((......)).)))))........(((((.((.....)).)))))....-- ( -36.50, z-score = -2.99, R) >droYak2.chr2L_random 1913381 111 - 4064425 GAUUCCGUAAGCUGCCCCGUUGCUGCGGGCUGCAGGUUACUUGCCGCUUUCCUGGCGGCUGUUCCACUUUCCUUCGGAGUCUGUGGCCGCUGCCGAUCACCAGCCACACUC ((((((((((.(((((((((....)))))..)))).))))..((((((.....))))))................))))))((((((.(.((.....)).).))))))... ( -46.80, z-score = -3.10, R) >droEre2.scaffold_4845 974651 111 - 22589142 GAGUCCGUAAGCUGCCUCGUUGCUGCGGGCUGCAGUUUACUUGCCGCUUUCCUGGCGGCUGUUCCACUUUCCUUCGGAGUCUGUGGCCGCUGCCGAUCACCAGCCACAUUC ((.(((((((((((((((((....)))))..)))))))))..((((((.....))))))................))).))((((((.(.((.....)).).))))))... ( -44.40, z-score = -2.64, R) >droAna3.scaffold_13417 6855903 111 - 6960332 GGGUCUGUAAUCUGCCCCCUUGCUUCGGGCUGCAGGUUACUUGCCGCUUUCCUGGCGGCUGUUCCACUUUCCUUCGGAGUCUGUGGCCGCUGCCGAUCACCAGCCACACUC .(((..(((((((((.(((.......)))..)))))))))..)))(((....(((((((.((..((..((((...))))..))..)).)))))))......)))....... ( -42.40, z-score = -2.21, R) >consensus GAAUCUGUAAUCUGCCCCGCUGCGGCAGGCUGCAGAGUACCUGCCGCUUUCCGACCGACUAAUCCACUUUCCUUCGAACUCUGAGGCCGCUGCAGAUCACCAGCCACACUC ((.((((((.((((((((((....)))))..))))).)))..(((((.......)))))................))).))...(((.(.((.....)).).)))...... (-28.31 = -25.90 + -2.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:43 2011