
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 925,344 – 925,516 |
| Length | 172 |
| Max. P | 0.749173 |

| Location | 925,344 – 925,454 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 65.95 |
| Shannon entropy | 0.56635 |
| G+C content | 0.45981 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.92 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
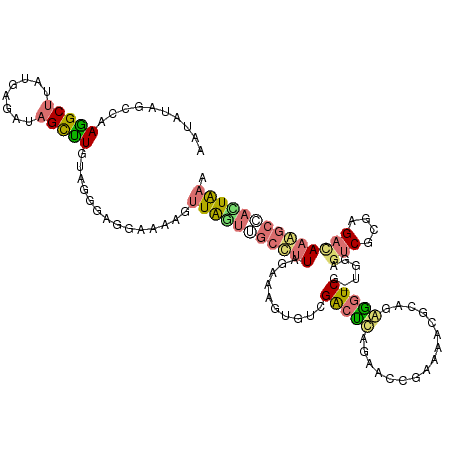
>dm3.chr2R 925344 110 - 21146708 AAUAUAGCCAAGGCUUAUGAGAUAGCUUG----GACGAAAGGUUAGUUGCCUUGGAAAGCGUCGACCCAGAUUCGAAUACGGAGAGGUC---GGAAGCGCGAGACAAAAGCACUGAC .......(((((.((........))))))----).(....)((((((.((.(((....(((((((((....((((....))))..))))---)...))))....)))..)))))))) ( -31.30, z-score = -1.82, R) >droSim1.chrU 5056859 117 - 15797150 AAUAUAGCCAAGGCUUAUGAGAUAGCCUGUAGGGAGGAAAAGUUAGUUGCCUUAGAAGGUGGCGGCUCAGAACCUAAAACUCAAGGGUCGGUAGAGUCGCGAGAUAAGGCUAUUAAA ...((((((..((((..((((....((....)).(((...((((.((..(((....)))..)))))).....)))....))))..))))......(((....)))..)))))).... ( -34.40, z-score = -1.56, R) >droSec1.super_165 59966 111 - 68140 AAUAUAGCAAAAGCGUACGAAAUAGCUUGCAGGGAAAGGAACCUGUUAGCCUUGGCG------GAAUCAGAUCGGAAAGCACAGGACUCAACGGAGUCAAAAGAUAAGGCUACUGAG ....((((....((...(((....(((.(((((........))))).))).((((..------...)))).)))....))....(((((....)))))..........))))..... ( -27.10, z-score = -1.31, R) >droYak2.chr2h_random 2814594 117 + 3774259 AAUAUAGCUAAGGCUUAUGAGAUAGUUUGUAGAGAGGAGAAGUUAACGGCUGUAGAAAGUGUCGGCUUAGAACCCAAAACGCAGAGGUCGUUGGAGUCGCGAGACAAAGCCAUUAAA ..((((((....)).)))).((((.(((.((.((.(..........)..)).)).))).))))(((((.....((((...((....))..)))).(((....))).)))))...... ( -27.10, z-score = -0.71, R) >consensus AAUAUAGCCAAGGCUUAUGAGAUAGCUUGUAGGGAGGAAAAGUUAGUUGCCUUAGAAAGUGUCGACUCAGAACCGAAAACGCAGAGGUCG_UGGAGUCGCGAGACAAAGCCACUAAA ..........(((((........)))))..............(((((((((((..........(((((................)))))......(((....)))))))))))))). (-14.04 = -14.92 + 0.88)

| Location | 925,418 – 925,516 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.39 |
| Shannon entropy | 0.47617 |
| G+C content | 0.43980 |
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -12.69 |
| Energy contribution | -14.12 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.588222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 925418 98 + 21146708 ----UUUCGUCCAAGCUAUCUCAUAAGCCUUGGCUAUAUUCAGGACCCC-AUGUUCCUUAUGGU------UAUCGGUCCCUUUAAGCACGAAAUAAAGGCGUUCAGCGC----------- ----(((((((((((((........)).))))).........(((((((-(((.....))))).------....)))))........)))))).....(((.....)))----------- ( -23.30, z-score = -1.23, R) >droSim1.chrU 5056936 120 + 15797150 UUUCCUCCCUACAGGCUAUCUCAUAAGCCUUGGCUAUAUUUGGAACCCCCAUGUUUCUUAUGGUUACACCUAUCGGUCCUUUUAGGGCUGAAAUAAAGGCGUUGAGCACCAUUUCUUCAU .............((....((((...((((((((((((...(((((......))))).))))))).......(((((((.....)))))))....)))))..))))..)).......... ( -31.30, z-score = -1.62, R) >droSec1.super_165 60037 120 + 68140 UCCUUUCCCUGCAAGCUAUUUCGUACGCUUUUGCUAUAUUCAGCCCCUCCAUAUUUCUAUUGGUCACCCCUAUUGGGCCCUUUAAUGCUGACACAAAAGCAUUUAGUGCCAUUUCUUCAU ..........(((.((......))..(((((((......(((((....(((.........)))....(((....))).........)))))..)))))))......)))........... ( -19.20, z-score = -0.64, R) >droYak2.chr2h_random 2814671 120 - 3774259 UCUCCUCUCUACAAACUAUCUCAUAAGCCUUAGCUAUAUUCGGAACCCCCAUGUUCCUUAUGGUCACCCCUAUUGGUCCCUUUAAGACCGAAAUAAAGGCGUUUAGCACCAUAUCCUCGU ..........................(((((.((((((...(((((......))))).))))))........((((((.......))))))....))))).................... ( -23.20, z-score = -2.05, R) >consensus UCUCCUCCCUACAAGCUAUCUCAUAAGCCUUGGCUAUAUUCAGAACCCCCAUGUUCCUUAUGGUCACCCCUAUCGGUCCCUUUAAGGCCGAAAUAAAGGCGUUUAGCACCAUUUCUUCAU ..........................(((((.((((((...(((((......))))).))))))........((((((.......))))))....))))).................... (-12.69 = -14.12 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:37 2011