
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,144,834 – 2,144,932 |
| Length | 98 |
| Max. P | 0.915415 |

| Location | 2,144,834 – 2,144,932 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 70.08 |
| Shannon entropy | 0.59817 |
| G+C content | 0.61385 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -14.90 |
| Energy contribution | -16.64 |
| Covariance contribution | 1.74 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
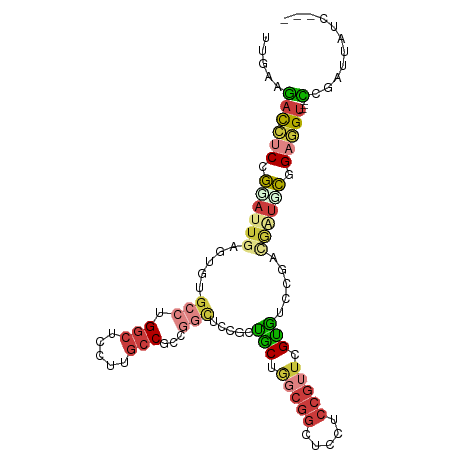
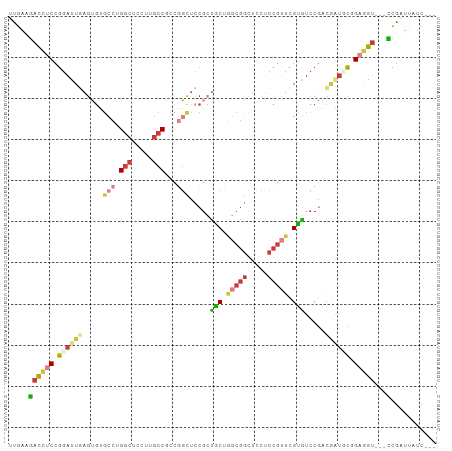
>dm3.chr2L 2144834 98 + 23011544 UUGAAGACCUCCGGAUUGAGUGUGCCUGGCUGAUUGCCGCCGGCUCCGCUACUGGCGGCUGCUCCGUUCGUGUCCGACGAUGCGGAGGU---CCGAUUAUC--- .....(((((((((((.(((((.(((.(((.....)))(((((........))))))))))))).))))(((((....)))))))))))---)........--- ( -43.60, z-score = -2.99, R) >droSim1.chr2L 2098569 98 + 22036055 UUGAAGACCUCCGGAUUGAGUGUGCCUGGCUUCUUGCCGCCGGCUCCGCUACUGGCGGCUGCUCCGUUCGUGUCCGACGAUUCGGAGGU---CCGAUUAUC--- .....(((((((((((((((((.(((.(((.....)))(((((........))))))))))))).((.((....)))))))))))))))---)........--- ( -42.70, z-score = -2.92, R) >droSec1.super_121 13117 98 + 59987 UUGAAGACCUCCGGAUUGAGUGUGCCUGGCUUCUUGCCGCCGGCUCCGCUACUGGCGGCUGCUCCGUUCGUGUCCGACAAUUCGGAGGU---CCGAUUAUC--- .....(((((((((((((((((.(((.(((.....)))(((((........))))))))))))).((.((....)))))))))))))))---)........--- ( -42.30, z-score = -3.18, R) >droYak2.chr2L 2116218 98 + 22324452 UUGAAGACCUCCGGAUUCAGUGUGCCGGGCUCCUUGCCACCGGCUGCGCUGCUGGCGGCUCCUCCGUUCGUGUCCGACGGUGCAGAGGU---UCGGUUUUC--- ..(((((((.((((...(((((((((((((.....))..))))).))))))))))..(..((((.((((((.....)))).)).)))).---.))))))))--- ( -38.50, z-score = -0.30, R) >droEre2.scaffold_4929 2179216 98 + 26641161 UUGAAGACCUCCGGAUUCAGUGUGCCAGGCUCCUUGCCGCCGGCUCCGCUGCUGGCGGCUCCUCCGUUCGUGUCCGACGAUGCGGAGGU---CCGGUUUUC--- ..(((((((...(((........(((.(((.....)))((((((......))))))))).(((((((((((.....)))).))))))))---)))))))))--- ( -47.00, z-score = -3.01, R) >droAna3.scaffold_12916 9850741 101 - 16180835 UUGAAGAUUUCCGGAUUGAGGGCGCCCGGCUCUUGGCCCACACUACCGGCGCCACCGCUGCCGCCGUUCGCGUCCUUUUCUCCAGCUGUGUUCCGGACUUC--- ..((((...((((((..((((((((..(((.....)))........(((((.((....)).)))))...))))))))..............))))))))))--- ( -35.19, z-score = -0.70, R) >droGri2.scaffold_15126 4660082 98 - 8399593 UUGAAUAUAUCAACGUUGAAUAAA---GUCUCUCCUCCCCCUGCUCC--UGCU-CCUGCUCCACCAUUCGCAGUCAUCGUGGUCGCUGCUGCAUCUCCCCCCAA ..((((...(((....))).....---...............((...--.)).-...........))))((((.((.((....)).))))))............ ( -11.30, z-score = 0.11, R) >consensus UUGAAGACCUCCGGAUUGAGUGUGCCUGGCUCCUUGCCGCCGGCUCCGCUGCUGGCGGCUCCUCCGUUCGUGUCCGACGAUGCGGAGGU___CCGAUUAUC___ ......(((((.((((((.....(((.(((.....)))...))).....(((.(((((.....))))).))).....)))))).)))))............... (-14.90 = -16.64 + 1.74)

| Location | 2,144,834 – 2,144,932 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 70.08 |
| Shannon entropy | 0.59817 |
| G+C content | 0.61385 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -17.70 |
| Energy contribution | -19.10 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
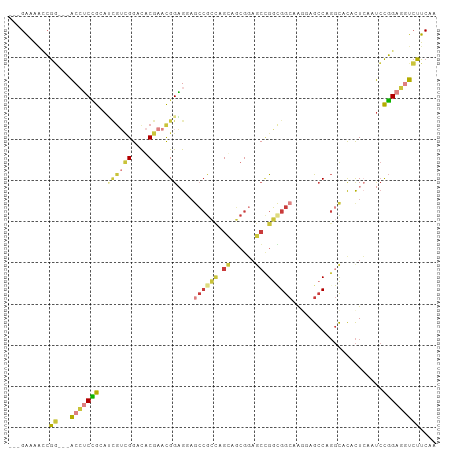
>dm3.chr2L 2144834 98 - 23011544 ---GAUAAUCGG---ACCUCCGCAUCGUCGGACACGAACGGAGCAGCCGCCAGUAGCGGAGCCGGCGGCAAUCAGCCAGGCACACUCAAUCCGGAGGUCUUCAA ---.......((---((((((((..(((((....)).)))..)).((((((.((......)).)))(((.....))).)))...........)))))))).... ( -39.80, z-score = -2.95, R) >droSim1.chr2L 2098569 98 - 22036055 ---GAUAAUCGG---ACCUCCGAAUCGUCGGACACGAACGGAGCAGCCGCCAGUAGCGGAGCCGGCGGCAAGAAGCCAGGCACACUCAAUCCGGAGGUCUUCAA ---.......((---(((((((...(((((....)).)))(((...((((.....)))).((((((........))).)))...)))....))))))))).... ( -39.50, z-score = -2.77, R) >droSec1.super_121 13117 98 - 59987 ---GAUAAUCGG---ACCUCCGAAUUGUCGGACACGAACGGAGCAGCCGCCAGUAGCGGAGCCGGCGGCAAGAAGCCAGGCACACUCAAUCCGGAGGUCUUCAA ---.......((---(((((((.(((.(((....)))...(((...((((.....)))).((((((........))).)))...)))))).))))))))).... ( -37.80, z-score = -2.38, R) >droYak2.chr2L 2116218 98 - 22324452 ---GAAAACCGA---ACCUCUGCACCGUCGGACACGAACGGAGGAGCCGCCAGCAGCGCAGCCGGUGGCAAGGAGCCCGGCACACUGAAUCCGGAGGUCUUCAA ---(((.(((..---.((((((.....(((....))).)))))).((((((.((......)).))))))..(((...(((....)))..)))...))).))).. ( -36.20, z-score = -0.69, R) >droEre2.scaffold_4929 2179216 98 - 26641161 ---GAAAACCGG---ACCUCCGCAUCGUCGGACACGAACGGAGGAGCCGCCAGCAGCGGAGCCGGCGGCAAGGAGCCUGGCACACUGAAUCCGGAGGUCUUCAA ---((...((((---(((((((..((((.....)))).)))))).((((((.((......)).)))(((.....))).)))........)))))...))..... ( -40.40, z-score = -1.46, R) >droAna3.scaffold_12916 9850741 101 + 16180835 ---GAAGUCCGGAACACAGCUGGAGAAAAGGACGCGAACGGCGGCAGCGGUGGCGCCGGUAGUGUGGGCCAAGAGCCGGGCGCCCUCAAUCCGGAAAUCUUCAA ---((((((((((.((....))(((......((((...(((((.((....)).)))))...))))(((((........))).)))))..))))))...)))).. ( -38.00, z-score = -0.12, R) >droGri2.scaffold_15126 4660082 98 + 8399593 UUGGGGGGAGAUGCAGCAGCGACCACGAUGACUGCGAAUGGUGGAGCAGG-AGCA--GGAGCAGGGGGAGGAGAGAC---UUUAUUCAACGUUGAUAUAUUCAA ......(((.(((((((.((..((((.((........)).)))).)).((-(((.--...))....((((......)---))).)))...)))).))).))).. ( -19.20, z-score = 0.27, R) >consensus ___GAAAACCGG___ACCUCCGCAUCGUCGGACACGAACGGAGGAGCCGCCAGCAGCGGAGCCGGCGGCAAGGAGCCAGGCACACUCAAUCCGGAGGUCUUCAA ..........((...(((((((..((((((....)).))))....((((((.((......)).))))))......................)))))))..)).. (-17.70 = -19.10 + 1.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:20 2011