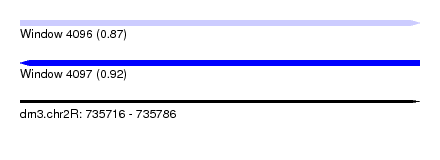
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 735,716 – 735,786 |
| Length | 70 |
| Max. P | 0.920564 |

| Location | 735,716 – 735,786 |
|---|---|
| Length | 70 |
| Sequences | 5 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 53.05 |
| Shannon entropy | 0.80763 |
| G+C content | 0.45340 |
| Mean single sequence MFE | -18.84 |
| Consensus MFE | -5.77 |
| Energy contribution | -5.57 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
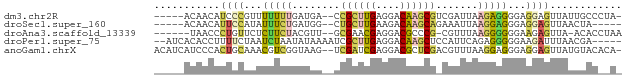
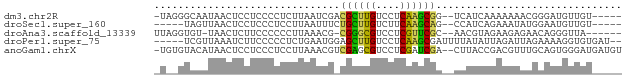
>dm3.chr2R 735716 70 + 21146708 -----ACAACAUCCCGUUUUUUUGAUGA--CCGCUUGAGGACAAGCGUCGAUUAAGAGGGGAGGAGUUAUUGCCCUA- -----......((((...((((((((((--(.(((((....)))))))).))))))))))))((.((....))))..- ( -20.10, z-score = -1.76, R) >droSec1.super_160 25237 66 - 40611 -----ACAACAUUCCAUAUUUCUGAUGG--CUGCUUGAAGACAAGCAGAAAUUAAGGAGGGAGGAGUUAACUA----- -----..(((.((((...((..((((..--(((((((....)))))))..))))..)).))))..))).....----- ( -20.30, z-score = -3.77, R) >droAna3.scaffold_13339 1902855 68 + 4599533 ------UAACCCUGUUCUCUUCUACGUU--GCGAACGAGGACGCCCG-CGUUUAAGGGGGGAAGAGUUA-ACACCUAA ------......((((((((((..(.((--..(((((.((....)).-))))).)).)..))))))..)-)))..... ( -19.10, z-score = -0.32, R) >droPer1.super_75 70162 71 + 290005 --AUCACACCUUUUCUAAUCUAAUAUAAAAUCGCUUGAGGACAAGCUCCAUUCAGAGGGGGAAGAUUUAACGA----- --......((((((((.((.....))......(((((....))))).......))))))))............----- ( -10.50, z-score = 0.26, R) >anoGam1.chrX 2449261 75 - 22145176 ACAUCAUCCCACUGCAAACGUCGGUAAG--UCGAUCGAGGACGCUCGACGUUUAAGGAGGGAGGAGUUAUGUACACA- ...((.((((.((..(((((((((...(--((.......)))..)))))))))..)).)))).))............- ( -24.20, z-score = -2.07, R) >consensus _____ACAACAUUCCAAAUUUCUGAUAA__CCGCUUGAGGACAAGCG_CGUUUAAGGGGGGAGGAGUUAACGAC__A_ ..................(((((........((((((....))))))........))))).................. ( -5.77 = -5.57 + -0.20)
| Location | 735,716 – 735,786 |
|---|---|
| Length | 70 |
| Sequences | 5 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 53.05 |
| Shannon entropy | 0.80763 |
| G+C content | 0.45340 |
| Mean single sequence MFE | -15.30 |
| Consensus MFE | -6.94 |
| Energy contribution | -5.86 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.83 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
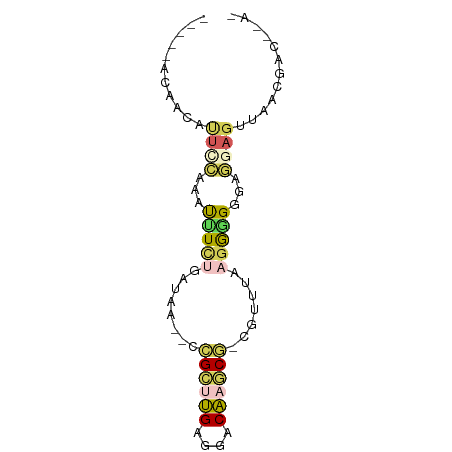
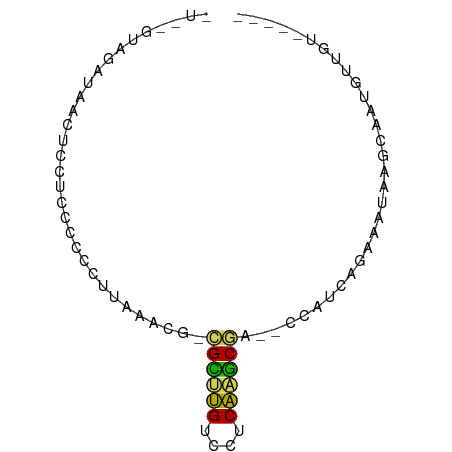
>dm3.chr2R 735716 70 - 21146708 -UAGGGCAAUAACUCCUCCCCUCUUAAUCGACGCUUGUCCUCAAGCGG--UCAUCAAAAAAACGGGAUGUUGU----- -....((((((.....((((.((......))((((((....)))))).--.............))))))))))----- ( -15.40, z-score = -1.03, R) >droSec1.super_160 25237 66 + 40611 -----UAGUUAACUCCUCCCUCCUUAAUUUCUGCUUGUCUUCAAGCAG--CCAUCAGAAAUAUGGAAUGUUGU----- -----....((((.................(((((((....)))))))--((((.(....)))))...)))).----- ( -12.90, z-score = -2.12, R) >droAna3.scaffold_13339 1902855 68 - 4599533 UUAGGUGU-UAACUCUUCCCCCCUUAAACG-CGGGCGUCCUCGUUCGC--AACGUAGAAGAGAACAGGGUUA------ .....(((-(..((((((..(........(-((((((....)))))))--...)..))))))))))......------ ( -15.80, z-score = 0.46, R) >droPer1.super_75 70162 71 - 290005 -----UCGUUAAAUCUUCCCCCUCUGAAUGGAGCUUGUCCUCAAGCGAUUUUAUAUUAGAUUAGAAAAGGUGUGAU-- -----..((((.(((((..(..(((((((((((((((....)))))...))))).)))))...)..))))).))))-- ( -11.80, z-score = -0.12, R) >anoGam1.chrX 2449261 75 + 22145176 -UGUGUACAUAACUCCUCCCUCCUUAAACGUCGAGCGUCCUCGAUCGA--CUUACCGACGUUUGCAGUGGGAUGAUGU -..........((((.((((.(..(((((((((...(((.......))--)....)))))))))..).)))).)).)) ( -20.60, z-score = -1.55, R) >consensus _U__GUAGAUAACUCCUCCCCCCUUAAACG_CGCUUGUCCUCAAGCGA__CCAUCAGAAAUAAGCAAUGUUGU_____ ...............................((((((....))))))............................... ( -6.94 = -5.86 + -1.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:29 2011