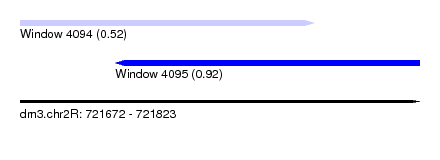
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 721,672 – 721,823 |
| Length | 151 |
| Max. P | 0.918960 |

| Location | 721,672 – 721,783 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 54.44 |
| Shannon entropy | 0.65863 |
| G+C content | 0.57183 |
| Mean single sequence MFE | -45.67 |
| Consensus MFE | -11.09 |
| Energy contribution | -12.77 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 721672 111 + 21146708 ----GGUACGGGCUAAAGCCCCAUUACCGCAACUUGGUCAGCCUGG-GGCC----UGACCCUAACCUUAAGGGGCUGAGCGGGGCCAGAACGAAGUGGUAUCUUCGUUACCUUCAGCAGG ----((((.((((....))))...))))((..((((.(((((((((-((..----...))))).((....)))))))).)))).....(((((((......))))))).......))... ( -45.00, z-score = -1.34, R) >droGri2.scaffold_15116 790869 120 + 1808639 GCCCAAAGAGGAAGGGAGAGCUCCUGCCACCAAGGGGUCAACCCUGUGGGCUGGAUGACCCAAACCGGUGAAAUCUCUGCGGUUCAAGGGUCAAGUGGUACCUCCGGUACCUCCAGCAGG ..((.....((..((((....)))).)).(((.((((....)))).)))((((((((((((.(((((..((....))..)))))...))))))...((((((...)))))))))))).)) ( -50.90, z-score = -1.47, R) >droVir3.scaffold_12823 181969 117 - 2474545 GUCAGAAACGCGAACCGAAUAUAACGUCACC---GGGACAACCCGGUGGCAAGGGUGAUCCAAACAGAAGAAACCUGAGCGGAUCGCGCACCAAAUGGUAUGUGCGCUACCUCAAAAAGG ........((((.((((........((((((---(((....)))))))))..(.(((((((...(((.......)))...))))))).)......))))...))))...(((.....))) ( -41.10, z-score = -3.28, R) >consensus G_C_GAAACGGGAAAAAGACCUAAUGCCACCA__GGGUCAACCCGGUGGCC_GG_UGACCCAAACCGAAGAAACCUGAGCGGAUCAAGAACCAAGUGGUAUCUCCGCUACCUCCAGCAGG .................(((((............((((((.((.........)).))))))....(((((....))))).)))))...........((((.......))))......... (-11.09 = -12.77 + 1.68)

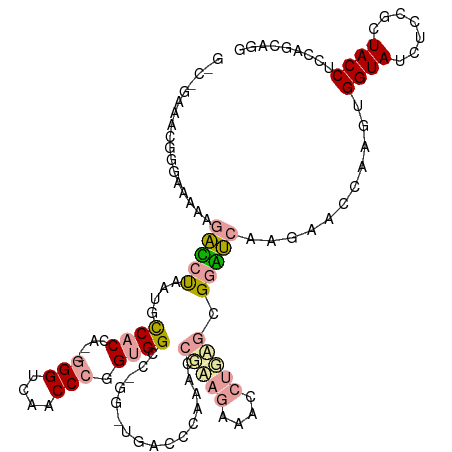
| Location | 721,708 – 721,823 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.17 |
| Shannon entropy | 0.59531 |
| G+C content | 0.57766 |
| Mean single sequence MFE | -48.00 |
| Consensus MFE | -20.01 |
| Energy contribution | -16.37 |
| Covariance contribution | -3.65 |
| Combinations/Pair | 1.74 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 721708 115 - 21146708 GGUGUCUUGGCCAGAGACAAAGCUUCUGCGGGUGACUUGCCCUGCUGAAGGUAACGAAGAUACCACUUCGUUCUGGCCCCGCUCAGCCCCUUAAGGUUAGGGUCAGGCCCCAGGC----- ((.((((.(((((((.......((((.((((((.....)))).)).))))...((((((......)))))))))))))..((((((((......)))).)))).)))).))....----- ( -43.80, z-score = -0.72, R) >droGri2.scaffold_15116 790909 120 - 1808639 GGCUCUUUGGCCUUUUUCAGGGCUUCAUCGGGGCUGAGUCCCUGCUGGAGGUACCGGAGGUACCACUUGACCCUUGAACCGCAGAGAUUUCACCGGUUUGGGUCAUCCAGCCCACAGGGU ((((((..(((((......))))).....))))))....((((((((((((((((...))))))...((((((..((((((..(......)..))))))))))))))))))....)))). ( -51.60, z-score = -1.92, R) >droVir3.scaffold_12823 182006 120 + 2474545 GGCUCCCUGGCCAUGCGCAAGGCUUCAUCGGGAUGAAAUCCCUUUUUGAGGUAGCGCACAUACCAUUUGGUGCGCGAUCCGCUCAGGUUUCUUCUGUUUGGAUCACCCUUGCCACCGGGU .....(((((...((.(((((((((((..(((((...)))))....)))))..((((((..........))))))((((((..((((.....))))..))))))..))))))))))))). ( -48.60, z-score = -2.81, R) >consensus GGCUCCUUGGCCAUACACAAGGCUUCAUCGGGACAAAAUCCCUGCUGGAGGUAACGAAGAUACCACUUGGUCCUCGAACCGCUCAGAUUUCUACGGUUUGGGUCACCCACCCCAC_GGGU (((......))).........((......(((((...))))).......((...(((.(((........))).)))..))))...(((((.........)))))................ (-20.01 = -16.37 + -3.65)

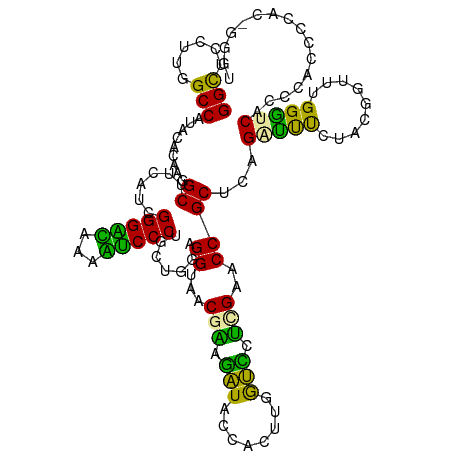
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:27 2011