| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,136,687 – 2,136,799 |
| Length | 112 |
| Max. P | 0.531925 |

| Location | 2,136,687 – 2,136,799 |
|---|---|
| Length | 112 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.19 |
| Shannon entropy | 0.71181 |
| G+C content | 0.44683 |
| Mean single sequence MFE | -23.01 |
| Consensus MFE | -11.16 |
| Energy contribution | -11.43 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.28 |
| Mean z-score | -0.39 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.531925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
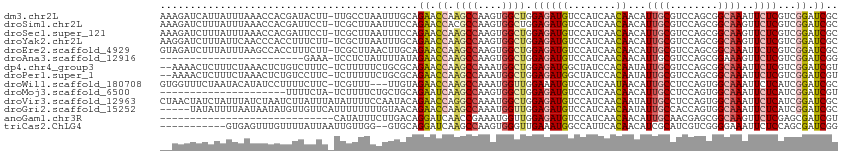
>dm3.chr2L 2136687 112 + 23011544 AAAGAUCAUUAUUUAAACCACGAUACUU-UUGCCUAAUUUGCAGAACCAAGCCAAGUGGCUGGAGAUGUCCAUCAACAACAUUGCGUCCAGCGGCAAAUUCUCGUCGGAUCGC .................((((((....(-(((((.....(((((..(((.(((....)))))).(((....))).......)))))......))))))...)))).))..... ( -27.00, z-score = -1.17, R) >droSim1.chr2L 2090128 112 + 22036055 AAAGAUCUUUAUUUAAACCACGAUUCCU-UCGCUUAAUUUCCAGAACCACGCCAAGUGGCUGGAGAUGUCCAUCAACAACAUUGCGUCCAGCGGCAAGUUCUCGUCGGAUCGC ...(((..............(((.....-)))....((((((((..((((.....))))))))))))....))).........((((((.((((.......)))).))).))) ( -26.20, z-score = -0.90, R) >droSec1.super_121 4972 112 + 59987 AAAGAUCUUUAUUUAAACCACGAUUCCU-UCGCUUAAUUUCCAGAACCAAGCCAAGUGGCUGGAGAUGUCCAUCAACAACAUUGCGUCCAGCGGCAAGUUCUCGUCGGAUCGC ...(((((............(((.....-)))..........(((((...(((.....(((((((((((.........)))))...)))))))))..)))))....))))).. ( -23.90, z-score = -0.24, R) >droYak2.chr2L 2108097 112 + 22324452 AAGGAUCUUUAUUCAACCCACCUUUCUU-UCGCUUAAUUUGCAGAACCAAGCCAAGUGGCUGGAGAUGUCCAUCAACAACAUUGCGUCCAGCGGCAAGUUCUCGUCGGAUCGC ..((((.(((.................(-((((.......)).)))(((.(((....))))))))).))))............((((((.((((.......)))).))).))) ( -24.50, z-score = 0.22, R) >droEre2.scaffold_4929 2171031 112 + 26641161 GUAGAUCUUUAUUUAAGCCACCUUUCUU-UCGCUUAACUUGCAGAACCAAGCCAAGUGGCUGGAGAUGUCCAUCAACAACAUUGCGUCCAGCGGCAAAUUCUCGUCGGAUCGC ...(((((........(((.........-.(((((..((((......))))..)))))(((((((((((.........)))))...)))))))))...........))))).. ( -26.71, z-score = -0.71, R) >droAna3.scaffold_12916 9842670 88 - 16180835 ------------------------GAAA-UCCUCUAUUUUAUAGAACCAAGCCAAGUGGCUGGAGAUGUCCAUCAACAACAUUGCGUCCAGCGGAAAGUUCUCGUCGGAUCGG ------------------------((.(-((.(((((...))))).(((.(((....)))))).))).))..............(((((.(((((....)).))).))).)). ( -19.10, z-score = 0.02, R) >dp4.chr4_group3 7386049 110 - 11692001 --AAAACUCUUUCUAAACUCUGUCUUUC-UCUUUUUCUGCGCAGAACCAAGCCAAAUGGCUGGAGAUGGCUAUCCACAAUAUUGCGUCCAGCGGCAAAUUCUCGUCGGAUCGU --...................(((..((-(((..((((....))))...((((....))))))))).))).............((((((.((((.......)))).))).))) ( -22.40, z-score = 0.12, R) >droPer1.super_1 4494670 110 - 10282868 --AAAACUCUUUCUAAACUCUGUCCUUC-UCUUUUUCUGCGCAGAACCAAGCCAAAUGGCUGGAGAUGGCUAUCCACAAUAUUGCGUCCAGCGGCAAAUUCUCGUCGGAUCGU --...................(((..((-(((..((((....))))...((((....))))))))).))).............((((((.((((.......)))).))).))) ( -22.30, z-score = 0.17, R) >droWil1.scaffold_180708 2068020 109 + 12563649 GUGGUUUCUAAUACAUAUCCUUUUCUUC-UCGUUU---UUGUAGAACCAAGCCAAAUGGUUGGAAAUGUCCAUCAAUAACAUUGCCUCCAGUGGCAAAUUCUCAUCGGAUCGC (((((((.....((((.(((.....(((-(((...---.)).))))...((((....))))))).))))............(((((......))))).........))))))) ( -20.70, z-score = -0.53, R) >droMoj3.scaffold_6500 29348168 91 - 32352404 ---------------------UUUUCUA-UCUUUUCUGCUGCAGAAUCAAGCCAAGUGGCUGGAGAUGUCCAUCAACAACAUUGCCUCCAGUGGCAAAUUCUCAUCGGAUCGU ---------------------.......-.....((((.((.(((((...((((..(((..((..((((.........))))..)).))).))))..))))))).)))).... ( -22.50, z-score = -0.85, R) >droVir3.scaffold_12963 7846790 113 + 20206255 CUAACUAUCUAUUUAUCUAAUCUUAUUUAUAUUUUCCAAUACAGAACCAGGCCAAAUGGCUGGAGAUGUCCAUCAACAAUAUUGCCUCCAGUGGCAAAUUCUCAUCGGAUCGC ...........................................((.((.((((....)))).(((((((......)))...(((((......)))))..))))...)).)).. ( -20.30, z-score = -0.59, R) >droGri2.scaffold_15252 10927556 108 + 17193109 -----UAUAUUUUAAUAAUAUGUUGUUCAUUUUUUUUGUAACAGAACCAAGCCAAAUGGUUGGAGAUGUCCAUCAACAAUAUUGCCACCAGUGGCAAAUUCUCAUCGGAUCGC -----((((((.....)))))).((((((.......)).))))((.((.((((....)))).(((((((......)))...((((((....))))))..))))...)).)).. ( -21.60, z-score = -0.76, R) >anoGam1.chr3R 50053666 84 - 53272125 -----------------------------CAUAUUUCUUGACAGGAUCAACCGAAAUGGUUGGAGAUGUCCAUCAACAACAUUGCAACGAGCGGCAAGUUCUCGAGCGAUCGU -----------------------------..........((((...((((((.....))))))...))))........(((((((...((((.....))))....))))).)) ( -17.60, z-score = 0.49, R) >triCas2.ChLG4 8264101 100 + 13894384 -----------GUGAGUUUGUUUUAUUAAUUGUUGG--GUGCAGGAUCAAGCCAAGUGGGUUGAAAUGGCCAUUCACAACAUCGCAUCGUCGGGGAAAUUCUCCAGCGAUCGG -----------(((((((((......)))......(--((.((...((((.((....)).))))..))))))))))).....((.(((((.((((.....)))).))))))). ( -27.30, z-score = -0.75, R) >consensus __AGAUCUUUAUUUAAACCAUGUUUCUU_UCGUUUAAUUUGCAGAACCAAGCCAAGUGGCUGGAGAUGUCCAUCAACAACAUUGCGUCCAGCGGCAAAUUCUCGUCGGAUCGC ...........................................((.((.((((....)))).((((((........))...((((........))))..))))...)).)).. (-11.16 = -11.43 + 0.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:18 2011