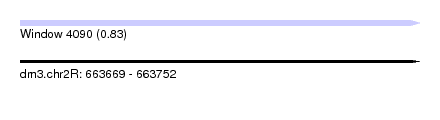
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 663,669 – 663,752 |
| Length | 83 |
| Max. P | 0.834764 |

| Location | 663,669 – 663,752 |
|---|---|
| Length | 83 |
| Sequences | 4 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 64.59 |
| Shannon entropy | 0.54375 |
| G+C content | 0.38899 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -7.31 |
| Energy contribution | -8.50 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 663669 83 + 21146708 CCGAUUAGUACCGAAAUC---ACUCAAAAAGGAAUUAACCGUUGCUCGCAGGCAACUUGAGUGUUUGAGUACACAAGUUUCUUAUA .......((((((((..(---((((((...((......))((((((....))))))))))))))))).)))).............. ( -22.70, z-score = -3.00, R) >droSim1.chr3h_random 181853 83 + 1452968 CCGAUUAGUAGCGAAAAC---ACUCAAAAUGGAAUUAACCGUUGCUCGCAGGCAACUUGAGUGUUUGAGUACAAAAGUUUCUUAUA .......(((.(..((((---((((((...((......))((((((....))))))))))))))))..)))).............. ( -22.90, z-score = -2.84, R) >droSec1.super_48 179566 72 - 230265 CCGAUCAGUAGCGAAAAA---ACUCAAAAAGGAAUUAACCGUUGCUCGCAGGCAACUUGAGUGUUUGAGUACAAA----------- .......(((.((((...---((((((...((......))((((((....)))))))))))).))))..)))...----------- ( -18.70, z-score = -2.26, R) >droVir3.scaffold_13561 100373 73 + 156932 -CCAACAGAAAUGAAGACCAAACCGAAAAUAAUAUUGAACUCUA--GGAGGGUAUUCCCAUAAUAAGACUACCAAA---------- -((..((....))..........(((........))).......--)).(((....))).................---------- ( -6.10, z-score = 0.75, R) >consensus CCGAUCAGUAGCGAAAAC___ACUCAAAAAGGAAUUAACCGUUGCUCGCAGGCAACUUGAGUGUUUGAGUACAAAA__________ .....................(((((((..((......))..((((((.((....)))))))))))))))................ ( -7.31 = -8.50 + 1.19)


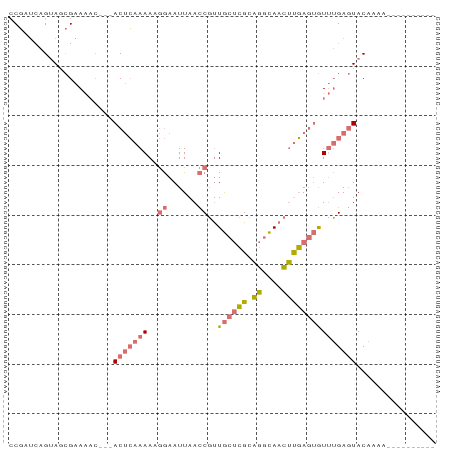
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:23 2011